4EDP
 
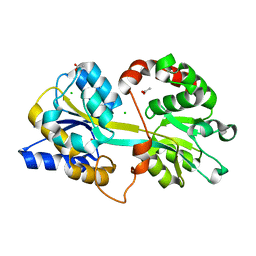 | | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124 | | Descriptor: | ABC transporter, substrate-binding protein, ACETATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-27 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124
To be Published
|
|
7TKV
 
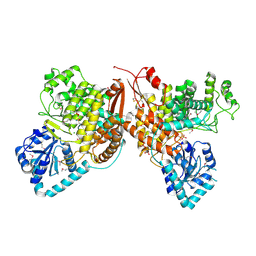 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-17 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
7THW
 
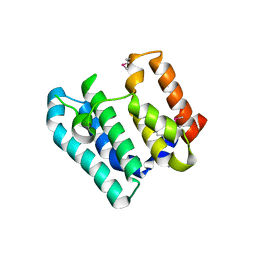 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | Authors: | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-12 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
7TJ1
 
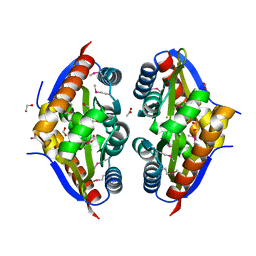 | | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Tesar, C, Pastore, T, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-14 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
To Be Published
|
|
7TMU
 
 | | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis
To Be Published
|
|
7TRW
 
 | | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | 3-HYDROXYBENZOIC ACID, LysR-family transcriptional regulatory protein, PHOSPHATE ION | | Authors: | Kim, Y, Tesar, C, Crawford, M, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7TRV
 
 | | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Crawford, M, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7TOC
 
 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | Authors: | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
7TVX
 
 | |
3H02
 
 | | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium. | | Descriptor: | BICARBONATE ION, Naphthoate synthase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3H0P
 
 | | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium. | | Descriptor: | S-malonyltransferase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7KPO
 
 | | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Response regulator, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae.
To Be Published
|
|
6CX8
 
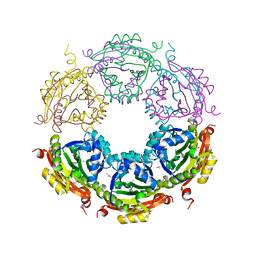 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, MANGANESE (II) ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions.
To Be Published
|
|
6D72
 
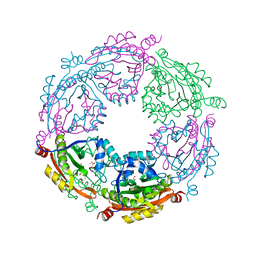 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions.
To Be Published
|
|
6DEB
 
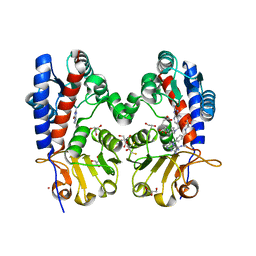 | | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional protein FolD, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni
To Be Published
|
|
7KOM
 
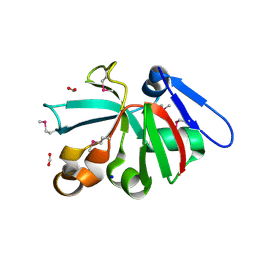 | | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6 | | Descriptor: | FORMIC ACID, Oxidored_molyb domain-containing protein, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6.
To Be Published
|
|
7KOS
 
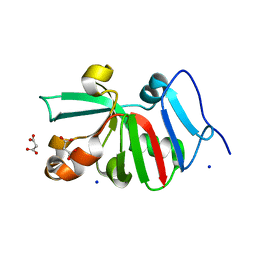 | | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | FORMIC ACID, MALONIC ACID, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7KP2
 
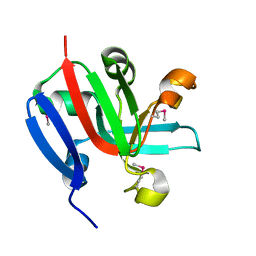 | | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin | | Descriptor: | L-NEOPTERIN, Putative Pterin Binding Protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Pshenychnyi, S, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin.
To Be Published
|
|
7KOU
 
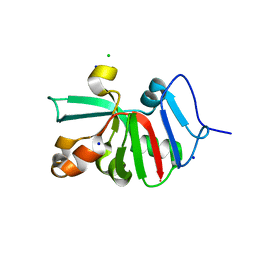 | | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7L6Y
 
 | |
7L6L
 
 | | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyribose operon repressor, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12.
To Be Published
|
|
7L75
 
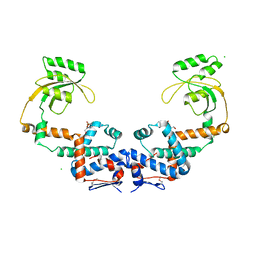 | | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Foldase protein PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans.
To Be Published
|
|
7L6Z
 
 | | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6
To Be Published
|
|
7L6J
 
 | | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative hydrolase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia
To Be Published
|
|
