6CN1
 
 | | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
6D0G
 
 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
6DB1
 
 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | Descriptor: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
6D72
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions.
To Be Published
|
|
6BK7
 
 | | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis | | Descriptor: | Elongation factor G, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis.
To be Published
|
|
6BI4
 
 | | 2.9 Angstrom Resolution Crystal Structure of dTDP-Glucose 4,6-dehydratase (rfbB) from Bacillus anthracis str. Ames in Complex with NAD. | | Descriptor: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-31 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-l-rhamnose biosynthetic pathway enzyme: dTDP-alpha-d-glucose 4,6-dehydratase, RfbB.
J.Struct.Biol., 202, 2018
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
6CX8
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, MANGANESE (II) ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions.
To Be Published
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6D0P
 
 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
6E1X
 
 | | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To Be Published
|
|
6E5Y
 
 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
6C4Q
 
 | | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Polyketide synthase Pks13 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To Be Published
|
|
6BXG
 
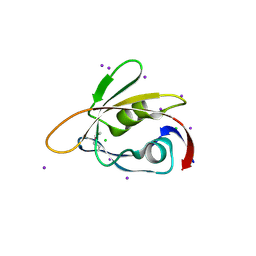 | | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide. | | Descriptor: | CHLORIDE ION, IODIDE ION, LEU-ILE-ALA, ... | | Authors: | Minasov, G, Shuvalova, L, Filippova, E.V, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide.
To Be Published
|
|
6C43
 
 | | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium. | | Descriptor: | Gamma-aminobutyraldehyde dehydrogenase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Tekleab, H, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-11 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.
To Be Published
|
|
6DAU
 
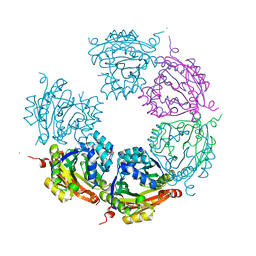 | | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae | | Descriptor: | GLYCEROL, Spermidine N1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Beahan, A, Kulyavtsev, P, Tan, L, Tran, D, Kuhn, M.L, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To be Published
|
|
5CNP
 
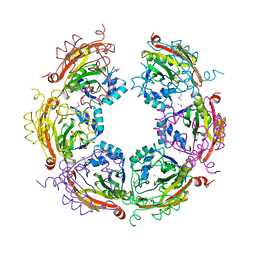 | | X-ray crystal structure of Spermidine n1-acetyltransferase from Vibrio cholerae. | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Osipiuk, J, VOLKART, L, MOY, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
5U2G
 
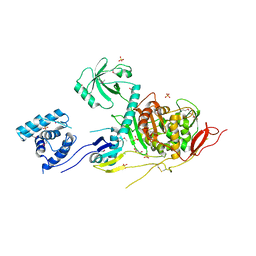 | | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae.
To Be Published
|
|
5U4O
 
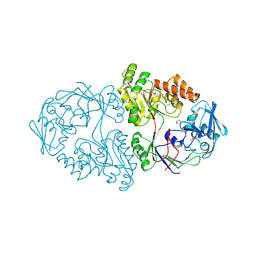 | | A 2.05A X-Ray Structureof A Bacterial Extracellular Solute-binding Protein, family 5 for Bacillus anthracis str. Ames | | Descriptor: | ABC transporter substrate-binding protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A 2.05A X-Ray Structureof A Bacterial Extracellular Solute-binding Protein, family 5 for Bacillus anthracis str. Ames
To Be Published
|
|
5U63
 
 | | Crystal structure of putative thioredoxin reductase from Haemophilus influenzae | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Michalska, K, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of putative thioredoxin reductase from Haemophilus influenzae
To Be Published
|
|
5TR3
 
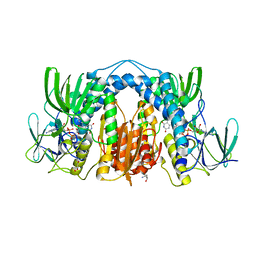 | | 2.5 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD.
To Be Published
|
|
5TR7
 
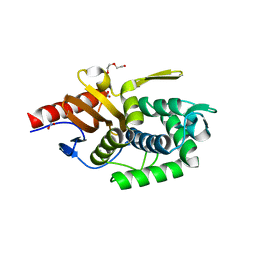 | | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961
To Be Published
|
|
5TSE
 
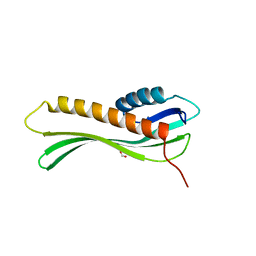 | | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii. | | Descriptor: | FORMIC ACID, LPS-assembly lipoprotein LptE | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii.
To Be Published
|
|
5U1O
 
 | | 2.3 Angstrom Resolution Crystal Structure of Glutathione Reductase from Vibrio parahaemolyticus in Complex with FAD. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3 Angstrom Resolution Crystal Structure of Glutathione Reductase from Vibrio parahaemolyticus in Complex with FAD.
To Be Published
|
|
5TTA
 
 | | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein | | Descriptor: | Putative exported protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Cordona-Correa, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein
To Be Published
|
|
