5RMD
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z57614330 | | Descriptor: | Helicase, N-ethyl-4-[(methylsulfonyl)amino]benzamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1429867185 | | Descriptor: | 4-methoxy-1H-indole, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLW
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z45705015 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z24758179 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RL7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z364321922 | | Descriptor: | 5-(acetylamino)-2-fluorobenzoic acid, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLN
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z364328788 | | Descriptor: | 3-(acetylamino)-4-fluorobenzoic acid, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z373768900 | | Descriptor: | Helicase, N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53860899 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLL
 
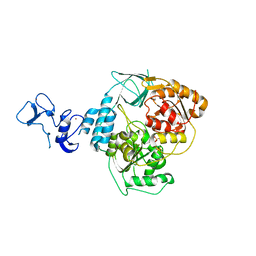 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z425387594 | | Descriptor: | 1-(2-ethoxyphenyl)piperazine, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLV
 
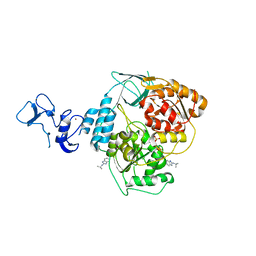 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z2467208649 | | Descriptor: | Helicase, N-(propan-2-yl)-1H-benzimidazol-2-amine, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM8
 
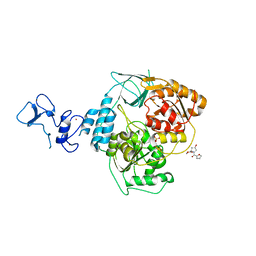 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1614545742 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLF
 
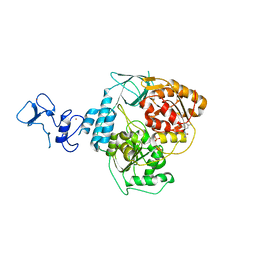 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z235341991 | | Descriptor: | Helicase, N-(2-methoxy-5-methylphenyl)glycinamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z744754722 | | Descriptor: | 2-(thiophen-2-yl)-1H-imidazole, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RME
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z26333434 | | Descriptor: | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5QPG
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000291a | | Descriptor: | 3-(azepan-1-ylmethyl)-1~{H}-indole, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPV
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000416a | | Descriptor: | (2~{S})-~{N}-(4-aminocarbonylphenyl)oxolane-2-carboxamide, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPM
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000500a | | Descriptor: | 1-(4-fluorophenyl)-3-(3-hydroxyphenyl)urea, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQB
 
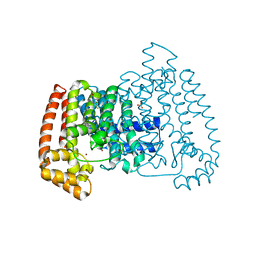 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000676a | | Descriptor: | 8-fluoranyl-5-methyl-1,2,3,6-tetrahydro-1,5-benzodiazocin-4-one, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPD
 
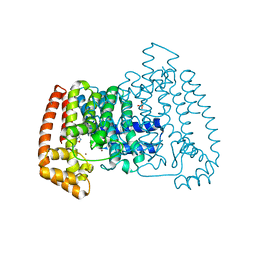 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000293a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPX
 
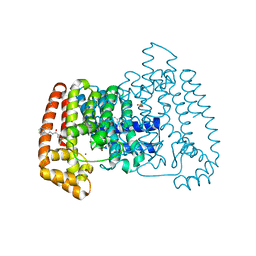 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000534a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPN
 
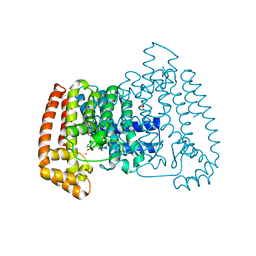 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000576a | | Descriptor: | 3-[(4-methylpiperidin-1-yl)methyl]-1H-indole, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ0
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with XST00000046b | | Descriptor: | 2-morpholin-4-ylaniline, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ6
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000530a | | Descriptor: | (3~{a}~{S},8~{a}~{S})-6-(phenylcarbonyl)-2,3,3~{a},7,8,8~{a}-hexahydropyrrolo[3,4-d]azepin-1-one, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPP
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000512a | | Descriptor: | 1-[2-(trifluoromethyloxy)phenyl]thiourea, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ2
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000693a | | Descriptor: | (2R)-1-{[(5-methylthiophen-2-yl)methyl]amino}propan-2-ol, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
