9GPB
 
 | |
2GPN
 
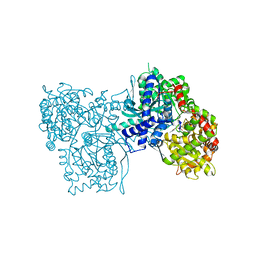 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
2K7G
 
 | | Solution Structure of varv F | | Descriptor: | Varv peptide F | | Authors: | Wang, C.K. | | Deposit date: | 2008-08-10 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Combined X-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold
J.Biol.Chem., 284, 2009
|
|
2JKZ
 
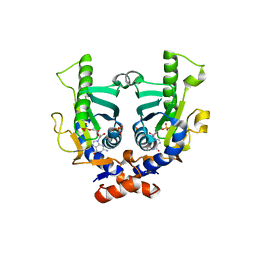 | | SACCHAROMYCES CEREVISIAE HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH GMP (GUANOSINE 5'- MONOPHOSPHATE) (ORTHORHOMBIC CRYSTAL FORM) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Moynie, L, Giraud, M.F, Breton, A, Boissier, F, Daignan-Fornier, B, Dautant, A. | | Deposit date: | 2008-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Protein Sci., 21, 2012
|
|
2JKY
 
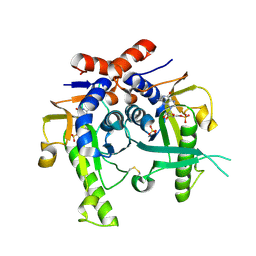 | | SACCHAROMYCES CEREVISIAE HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH GMP (GUANOSINE 5'- MONOPHOSPHATE) (TETRAGONAL CRYSTAL FORM) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Moynie, L, Giraud, M.F, Breton, A, Boissier, F, Daignan-Fornier, B, Dautant, A. | | Deposit date: | 2008-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Protein Sci., 21, 2012
|
|
2MBT
 
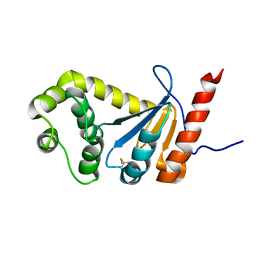 | | NMR study of PaDsbA | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Rimmer, K, Mohanty, B, Scanlon, M.J. | | Deposit date: | 2013-08-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
2AMV
 
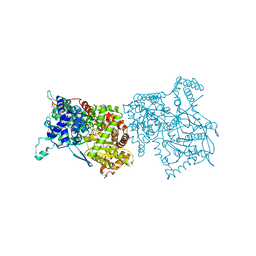 | | THE STRUCTURE OF GLYCOGEN PHOSPHORYLASE B WITH AN ALKYL-DIHYDROPYRIDINE-DICARBOXYLIC ACID | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Zographos, S.E, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor.
Structure, 5, 1997
|
|
4DM3
 
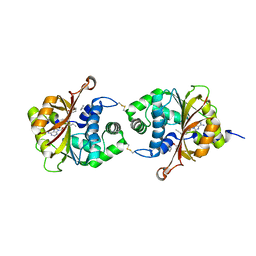 | | Crystal structure of human PNMT in complex adohcy, resorcinol and imidazole | | Descriptor: | IMIDAZOLE, Phenylethanolamine N-methyltransferase, RESORCINOL, ... | | Authors: | Nair, P.C, Malde, A.K, Mark, A.E. | | Deposit date: | 2012-02-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4001 Å) | | Cite: | Missing fragments: detecting cooperative binding in fragment-based drug design.
ACS Med Chem Lett, 3, 2012
|
|
2PRI
 
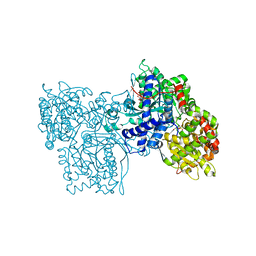 | | BINDING OF 2-DEOXY-GLUCOSE-6-PHOSPHATE TO GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLYCOGEN PHOSPHORYLASE B, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Zographos, S.E, Johnson, L.N, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The binding of 2-deoxy-D-glucose 6-phosphate to glycogen phosphorylase b: kinetic and crystallographic studies.
J.Mol.Biol., 254, 1995
|
|
1GPA
 
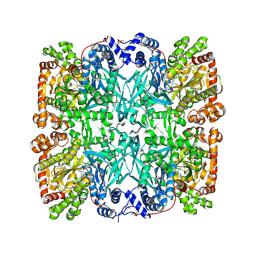 | |
1A8I
 
 | | SPIROHYDANTOIN INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | BETA-D-GLUCOPYRANOSE SPIROHYDANTOIN, GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-25 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
1GPB
 
 | |
1B4D
 
 | | AMIDOCARBAMATE INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE B), ... | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1998-12-18 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
7GPB
 
 | | STRUCTURAL MECHANISM FOR GLYCOGEN PHOSPHORYLASE CONTROL BY PHOSPHORYLATION AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE B, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Barford, D, Hu, S.-H, Johnson, L.N. | | Deposit date: | 1990-11-13 | | Release date: | 1992-10-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP.
J.Mol.Biol., 218, 1991
|
|
8RLJ
 
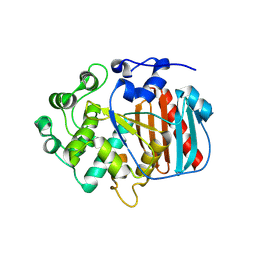 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | Class C beta-lactamase-related serine hydrolase | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
8RLK
 
 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Class C beta-lactamase-related serine hydrolase, MAGNESIUM ION, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
8RLL
 
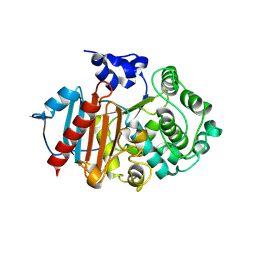 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | Class C beta-lactamase-related serine hydrolase | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
7S1C
 
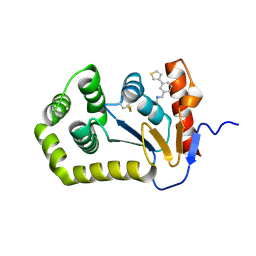 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001897 (compound 1) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-3-ylphenyl)methanamine | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Sharma, P. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1L
 
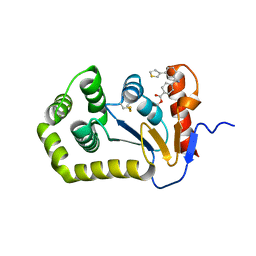 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001896 (compound 72) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, methyl cis-4-({[3-(thiophen-3-yl)benzyl]amino}methyl)cyclohexanecarboxylate | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1D
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001877 (compound 39) | | Descriptor: | 1-[3-(thiophen-3-yl)benzyl]piperidin-2-one, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1F
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001886 (compound 38) | | Descriptor: | 1-[(3-thiophen-3-ylphenyl)methyl]-3~{H}-pyrrol-2-one, COPPER (II) ION, GLYCEROL, ... | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
5QKO
 
 | |
5QL3
 
 | |
5QLM
 
 | |
5QM0
 
 | |
