7C3J
 
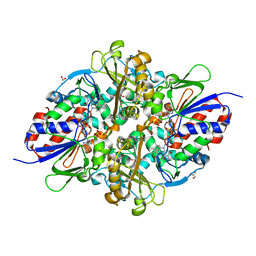 | | Structure of L-lysine oxidase D212A/D315A in complex with L-phenylalanine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
2EYZ
 
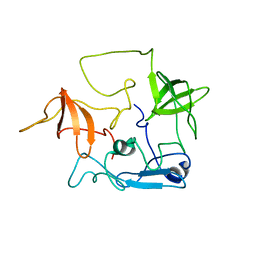 | | CT10-Regulated Kinase isoform II | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYW
 
 | |
4DGU
 
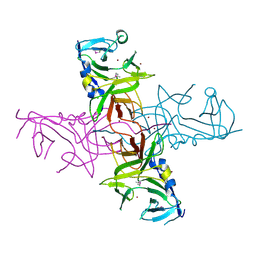 | |
2EYY
 
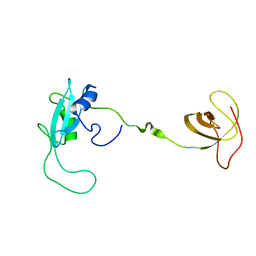 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3DGP
 
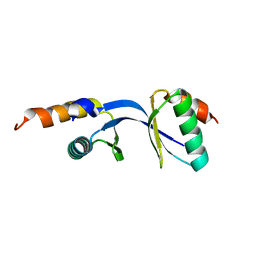 | | Crystal Structure of the complex between Tfb5 and the C-terminal domain of Tfb2 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, RNA polymerase II transcription factor B subunit 5 | | Authors: | Kainov, D.E, Cavarelli, J, Egly, J.M, Poterszman, A. | | Deposit date: | 2008-06-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for group A trichothiodystrophy
Nat.Struct.Mol.Biol., 15, 2008
|
|
4H40
 
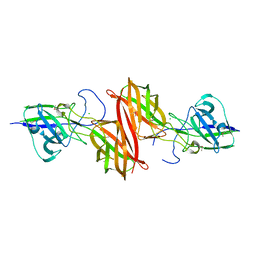 | |
1RAX
 
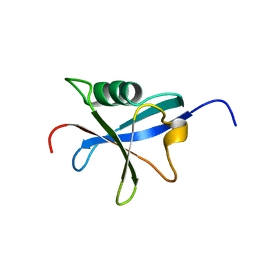 | |
4GPV
 
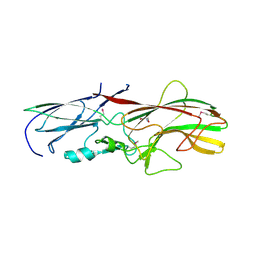 | |
2EYX
 
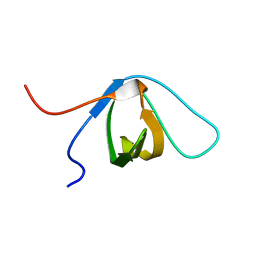 | |
2EYV
 
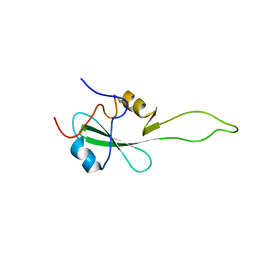 | | SH2 domain of CT10-Regulated Kinase | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7Y7D
 
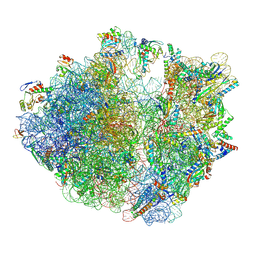 | | Structure of the Bacterial Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7C
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(G34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7F
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(ManQ34) and mRNA(GAC) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7E
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7G
 
 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7H
 
 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAC) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
4QDG
 
 | |
4Q98
 
 | |
4QB7
 
 | |
2IO5
 
 | | Crystal structure of the CIA- histone H3-H4 complex | | Descriptor: | ASF1A protein, Histone H3.1, Histone H4 | | Authors: | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | Deposit date: | 2006-10-10 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
3GF8
 
 | |
7LTJ
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with a non-covalent inhibitor Mcule-5948770040 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-02-19 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Noncovalent Inhibitor.
J.Chem.Inf.Model., 62, 2022
|
|
8SJQ
 
 | | [3T16] Self-assembling right-handed tensegrity triangle with 16 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*GP*CP*CP*TP*GP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.19 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJM
 
 | | [3T12] Self-assembling left-handed tensegrity triangle with 12 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*AP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*TP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
