2NQU
 
 | | MoeA E188Q | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQQ
 
 | | MoeA R137Q | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2Q5W
 
 | |
2QIE
 
 | | Staphylococcus aureus molybdopterin synthase in complex with precursor Z | | Descriptor: | (2R,4AR,5AR,11AR,12AS)-8-AMINO-2-HYDROXY-4A,5A,9,11,11A,12A-HEXAHYDRO[1,3,2]DIOXAPHOSPHININO[4',5':5,6]PYRANO[3,2-G]PTERIDINE-10,12(4H,6H)-DIONE 2-OXIDE, Molybdopterin synthase small subunit, Molybdopterin-converting factor subunit 2 | | Authors: | Daniels, J.N, Schindelin, H. | | Deposit date: | 2007-07-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a molybdopterin synthase-precursor Z complex: insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency.
Biochemistry, 47, 2008
|
|
2FTS
 
 | |
1IX6
 
 | | Aspartate Aminotransferase Active Site Mutant V39F | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
2G9G
 
 | | Crystal structure of His-tagged mouse PNGase C-terminal domain | | Descriptor: | GLYCEROL, SULFATE ION, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FU3
 
 | |
2G9F
 
 | | Crystal structure of intein-tagged mouse PNGase C-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HBA
 
 | | Crystal Structure of N-terminal Domain of Ribosomal Protein L9 (NTL9) K12M | | Descriptor: | 50S ribosomal protein L9, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2HVF
 
 | | Crystal Structure of N-terminal Domain of Ribosomal Protein L9 (NTL9), G34dA | | Descriptor: | 50S ribosomal protein L9, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Anil, B, Kim, E.Y, Cho, J.H, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-07-28 | | Release date: | 2007-06-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Detecting and quantifying strain in protein folding
To be Published
|
|
2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | Descriptor: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | Authors: | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
1IX8
 
 | | Aspartate Aminotransferase Active Site Mutant V39F/N194A | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX7
 
 | | Aspartate Aminotransferase Active Site Mutant V39F maleate complex | | Descriptor: | Aspartate Aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
2HPL
 
 | | Crystal structure of the mouse p97/PNGase complex | | Descriptor: | C-terminal of mouse p97/VCP, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HPJ
 
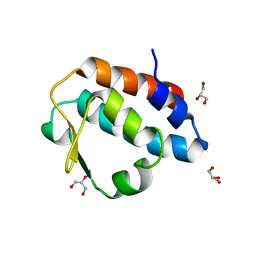 | | Crystal structure of the PUB domain of mouse PNGase | | Descriptor: | GLYCEROL, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I74
 
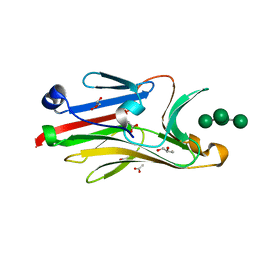 | | Crystal structure of mouse Peptide N-Glycanase C-terminal domain in complex with mannopentaose | | Descriptor: | ACETATE ION, GLYCEROL, PNGase, ... | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7ZH9
 
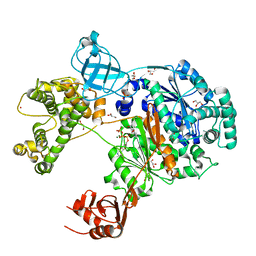 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7ZJP
 
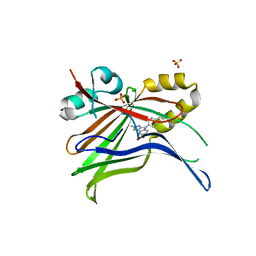 | | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 | | Descriptor: | 2-methyl-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxylic acid, SULFATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Freire, F, Heinrich, T, Petersson, C, Schneider, R, Garg, S, Schwarz, D, Gunera, J, Seshire, A, Koetzner, L, Schlesiger, S, Musil, D, Schilke, H, Doerfel, B, Diehl, P, Boepple, P, Lemos, A.R, Sousa, P.M.F, Freire, F, Bandeiras, T.M, Carswell, E, Pearson, N, Sirohi, S, Hooker, M, Trivier, E, Broome, R, Balsiger, A, Crowden, A, Dillon, C, Wienke, D. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 .
J.Med.Chem., 65, 2022
|
|
7ZTL
 
 | |
8A1I
 
 | |
6TNB
 
 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 41 | | Descriptor: | (2~{R})-2-(4-fluorophenyl)-~{N}-[4-[2-[(2-methoxy-4-methylsulfonyl-phenyl)amino]-[1,2,4]triazolo[1,5-a]pyridin-6-yl]phenyl]propanamide, CHLORIDE ION, Dual specificity protein kinase TTK | | Authors: | Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Marquardt, T, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
6TNC
 
 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 46 | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Marquardt, T, Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
6TN9
 
 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 16 | | Descriptor: | Dual specificity protein kinase TTK, [4-[[6-(3,5-dimethyl-4-oxidanyl-phenyl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]amino]phenyl]-morpholin-4-yl-methanone | | Authors: | Marquardt, T, Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
6TND
 
 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 79 | | Descriptor: | BAY 1217389, Dual specificity protein kinase TTK | | Authors: | Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Marquardt, T, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
