3UH3
 
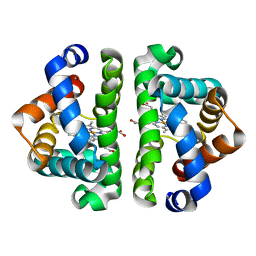 | | HBI (L36V) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UHB
 
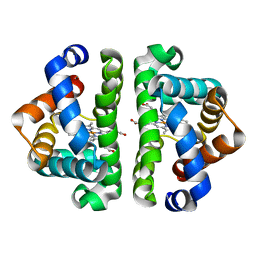 | | HBI (R104K) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UHZ
 
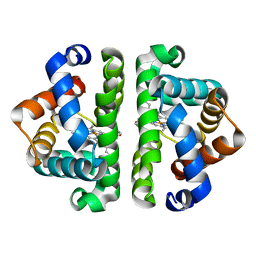 | | HBI (T72A) deoxy | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GD8
 
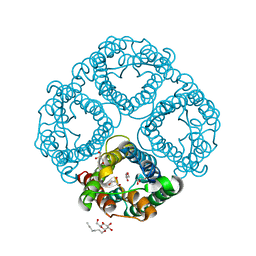 | | Crystal Structure of Human Aquaporin 4 at 1.8 and its Mechanism of Conductance | | Descriptor: | Aquaporin-4, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Ho, J.D, Yeh, R, Sandstrom, A, Chorny, I, Harries, W.E.C, Robbins, R.A, Miercke, L.J.W, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1EY6
 
 | |
1EYD
 
 | |
1EY0
 
 | |
1DUY
 
 | | CRYSTAL STRUCTURE OF HLA-A*0201/OCTAMERIC TAX PEPTIDE COMPLEX | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A2*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1EY4
 
 | |
1EYA
 
 | |
1EY5
 
 | |
1EY9
 
 | |
1F6B
 
 | | CRYSTAL STRUCTURE OF SAR1-GDP COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SAR1, ... | | Authors: | Huang, M, Wilson, I.A, Balch, W.E. | | Deposit date: | 2000-06-21 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export.
J.Cell Biol., 155, 2001
|
|
1EZ8
 
 | |
1EZ6
 
 | |
1EYC
 
 | |
1FMU
 
 | | STRUCTURE OF NATIVE PROTEINASE A IN P3221 SPACE GROUP. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1D5T
 
 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, SULFATE ION | | Authors: | Peng, L, Zeng, K, Heine, A, Moyer, B, Greasley, S.E, Kuhn, P, Balch, W.E, Wilson, I.A. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A new functional domain of guanine nucleotide dissociation inhibitor (alpha-GDI) involved in Rab recycling.
Traffic, 1, 2000
|
|
1FMX
 
 | | STRUCTURE OF NATIVE PROTEINASE A IN THE SPACE GROUP P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1DPJ
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1DP5
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT INHIBITOR | | Descriptor: | PROTEINASE A, PROTEINASE INHIBITOR IA3, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-23 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1DUZ
 
 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A 0201) IN COMPLEX WITH A NONAMERIC PEPTIDE FROM HTLV-1 TAX PROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1EY8
 
 | |
1EY7
 
 | |
1CDL
 
 | |
