4IIG
 
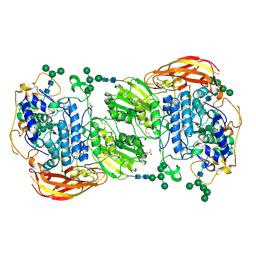 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with D-glucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIE
 
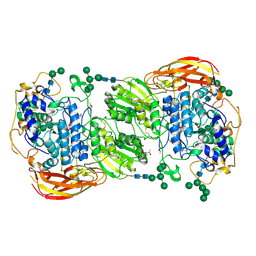 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with calystegine B(2) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
6IES
 
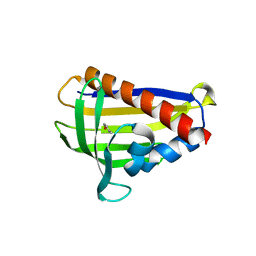 | | Onion lachrymatory factor synthase (LFS) containing (E)-2-propen 1-ol (crotyl alcohol) | | Descriptor: | (2E)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Sato, Y, Arakawa, T, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase.
Acs Catalysis, 10, 2020
|
|
3VJ7
 
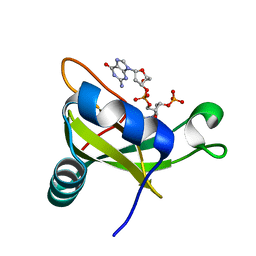 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 R33Q mutant | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Fushinobu, S, Ogawa, T, Hidaka, M, Masaki, H. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5
J.Biochem., 152, 2012
|
|
1J1I
 
 | | Crystal structure of a His-tagged Serine Hydrolase Involved in the Carbazole Degradation (CarC enzyme) | | Descriptor: | meta cleavage compound hydrolase | | Authors: | Habe, H, Morii, K, Fushinobu, S, Nam, J.W, Ayabe, Y, Yoshida, T, Wakagi, T, Yamane, H, Nojiri, H, Omori, T. | | Deposit date: | 2002-12-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme).
Biochem.Biophys.Res.Commun., 303, 2003
|
|
7CL7
 
 | |
7CL9
 
 | |
3WIQ
 
 | | Crystal structure of kojibiose phosphorylase complexed with kojibiose | | Descriptor: | Kojibiose phosphorylase, SULFATE ION, alpha-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
3WIR
 
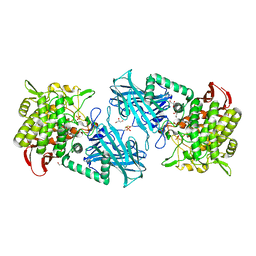 | | Crystal structure of kojibiose phosphorylase complexed with glucose | | Descriptor: | GLYCEROL, Kojibiose phosphorylase, PHOSPHATE ION, ... | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
6JU1
 
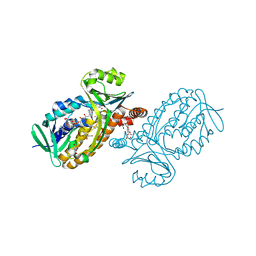 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
6KPO
 
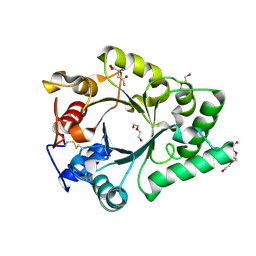 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine-Asn | | Descriptor: | ASPARAGINE, Chitinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPN
 
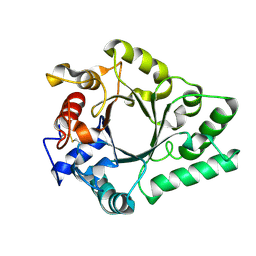 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine | | Descriptor: | Chitinase, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPM
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
2Z8F
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-tetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, SODIUM ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
2Z8D
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
2Z8E
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with galacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
3AC0
 
 | | Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose | | Descriptor: | Beta-glucosidase I, beta-D-glucopyranose | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
3ACT
 
 | |
3AHG
 
 | | H64A mutant of Phosphoketolase from Bifidobacterium Breve complexed with a tricyclic ring form of thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[(9aR)-2,7-dimethyl-9a,10-dihydro-5H-pyrimido[4,5-d][1,3]thiazolo[3,2-a]pyrimidin-8-yl]ethyl trihydrogen diphosphate, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHI
 
 | | H320A mutant of Phosphoketolase from Bifidobacterium Breve complexed with acetyl thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHD
 
 | | Phosphoketolase from Bifidobacterium Breve complexed with 2-acetyl-thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHJ
 
 | | H553A mutant of Phosphoketolase from Bifidobacterium Breve | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3ACS
 
 | |
3AHC
 
 | | Resting form of Phosphoketolase from Bifidobacterium Breve | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
