3NLL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57A OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-10 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4NUL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: D58P OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4NLL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57D OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
5NUL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57T SEMIQUINONE (150K) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-20 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4D7U
 
 | | The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa | | Descriptor: | COPPER (II) ION, ENDOGLUCANASE II, GLYCEROL | | Authors: | Borisova, A.S, Isaksen, T, Mathiesen, G, Sorlie, M, Sandgren, M, Eijsink, V.G.H, Dimarogona, M. | | Deposit date: | 2014-11-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity
J.Biol.Chem., 290, 2015
|
|
4D7V
 
 | | The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa | | Descriptor: | ACETATE ION, ENDOGLUCANASE II, GLYCEROL, ... | | Authors: | Borisova, A.S, Isaksen, T, Sandgren, M, Sorlie, M, Eijsink, V.G.H, Dimarogona, M. | | Deposit date: | 2014-11-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity
J.Biol.Chem., 290, 2015
|
|
4I8D
 
 | | Crystal Structure of Beta-D-glucoside glucohydrolase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucoside glucohydrolase, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Helmich, K.E, Banerjee, G, Bianchetti, C.M, Gudmundsson, M, Sandgren, M, Walton, J.D, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 beta-Glucosidase, Cel3A from Hypocrea jecorina.
J.Biol.Chem., 289, 2014
|
|
5OA5
 
 | | CELLOBIOHYDROLASE I (CEL7A) FROM HYPOCREA JECORINA WITH IMPROVED THERMAL STABILITY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exoglucanase 1, GLYCEROL | | Authors: | Goedegebuur, F, Hansson, H, Karkehabadi, S, Mikkelsen, N, Stahlberg, J, Sandgren, M. | | Deposit date: | 2017-06-20 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving the thermal stability of cellobiohydrolase Cel7A from Hypocrea jecorina by directed evolution.
J. Biol. Chem., 292, 2017
|
|
4PI9
 
 | | Crystal structure of S. Aureus Autolysin E in complex with muropeptide NAM-L-ALA-D-iGLU | | Descriptor: | (4R)-4-[[(2S)-2-[[(2R)-2-[(2R,3S,4R,5R,6R)-5-acetamido-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]oxypropanoyl]amino]propanoyl]amino]-5-azanyl-5-oxidanylidene-pentanoic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers.
IUCrJ, 4, 2017
|
|
4PI7
 
 | | Crystal structure of S. Aureus Autolysin E in complex with disaccharide NAM-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4PIA
 
 | | Crystal structure of S. Aureus Autolysin E | | Descriptor: | Autolysin E, CHLORIDE ION | | Authors: | Mihelic, M, Renko, M, Dobersek, A, Bedrac, L, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4PI8
 
 | | Crystal structure of catalytic mutant E138A of S. Aureus Autolysin E in complex with disaccharide NAG-NAM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
1NK3
 
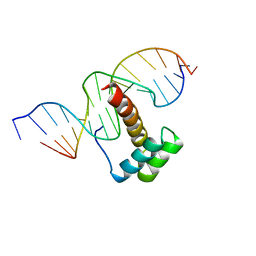 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NK2
 
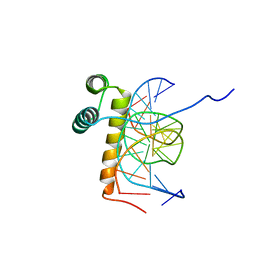 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
6HVO
 
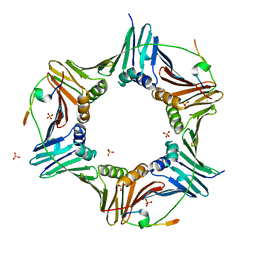 | | Crystal structure of human PCNA in complex with three peptides of p12 subunit of human polymerase delta | | Descriptor: | DNA polymerase delta subunit 4, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Gonzalez-Magana, A, Romano-Moreno, M, Rojas, A.L, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The p12 subunit of human polymerase delta uses an atypical PIP box for molecular recognition of proliferating cell nuclear antigen (PCNA).
J.Biol.Chem., 294, 2019
|
|
1ZN0
 
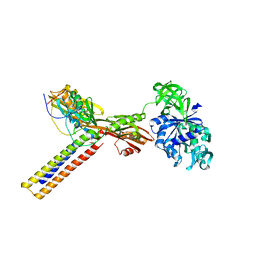 | | Coordinates of RRF and EF-G fitted into Cryo-EM map of the 50S subunit bound with both EF-G (GDPNP) and RRF | | Descriptor: | 16S RIBOSOMAL RNA, ELONGATION FACTOR G, Ribosome recycling factor | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
4DUH
 
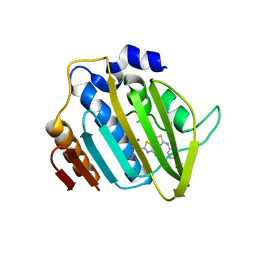 | | Crystal structure of 24 kDa domain of E. coli DNA gyrase B in complex with small molecule inhibitor | | Descriptor: | 4-{[4'-methyl-2'-(propanoylamino)-4,5'-bi-1,3-thiazol-2-yl]amino}benzoic acid, DNA gyrase subunit B | | Authors: | Brvar, M, Renko, M, Perdih, A, Solmajer, T, Turk, D. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of substituted 4,5'-bithiazoles as novel DNA gyrase inhibitors.
J.Med.Chem., 55, 2012
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
4ALE
 
 | | Structure changes of Polysaccharide monooxygenase CBM33A from Enterococcus faecalis by X-ray induced photoreduction. | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALQ
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALC
 
 | | X-Ray photoreduction of Polysaccharide monooxigenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
1ANX
 
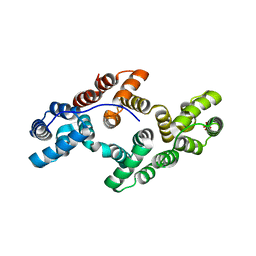 | |
1A4P
 
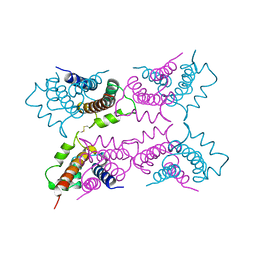 | | P11 (S100A10), LIGAND OF ANNEXIN II | | Descriptor: | S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
4ALS
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALT
 
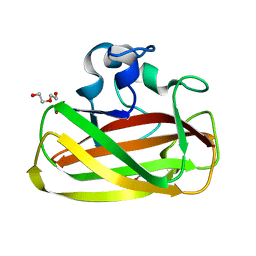 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
