4G12
 
 | | Crystal structure of putative TetR family transcriptional regulator, Fad35R, from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Probable transcriptional regulatory protein (Probably TETR-FAMILY) | | Authors: | Singh, A.K, Manjasetty, B.A, Singh, V, Mittal, M, Kumaran, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-07-10 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal structure of putative TetR family transcriptional regulator, Fad35R, from Mycobacterium tuberculosis
to be published
|
|
4GXA
 
 | |
4LI3
 
 | |
2ISQ
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Arabidopsis Thaliana in Complex with C-Terminal Peptide from Arabidopsis Serine Acetyltransferase | | Descriptor: | Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Francois, J.A, Kumaran, S, Jez, J.M. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for interaction of o-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex.
Plant Cell, 18, 2006
|
|
4LQ5
 
 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, O-acetylserine at 2.8A | | Descriptor: | HTH-type transcriptional regulator CysB, O-ACETYLSERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | rystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, O-acetylserine at 2.8A
TO BE PUBLISHED
|
|
4LQ2
 
 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium in complex with effector ligand, O-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, O-ACETYLSERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium in complex with effector ligand, O-acetylserine
TO BE PUBLISHED
|
|
4NU8
 
 | |
4LON
 
 | |
4M4G
 
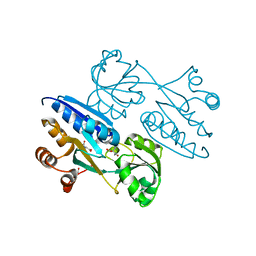 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, N-acetylserine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, N-acetylserine
To be Published
|
|
4ORE
 
 | |
2PBQ
 
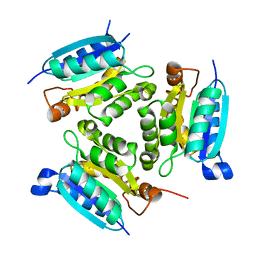 | | Crystal structure of molybdenum cofactor biosynthesis (aq_061) From aquifex aeolicus VF5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of molybdenum cofactor biosynthesis (aq_061) from aquifex aeolicus VF5
to be published
|
|
2QQ1
 
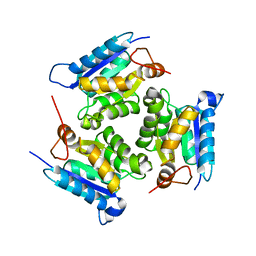 | | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5
To be Published
|
|
5XCW
 
 | |
5XCP
 
 | |
5Z3I
 
 | |
5Z3J
 
 | |
5Z37
 
 | |
5XCN
 
 | |
5XA2
 
 | |
5Y9B
 
 | | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with DON | | Descriptor: | 1,2-ETHANEDIOL, 6-DIAZENYL-5-OXO-L-NORLEUCINE, CALCIUM ION, ... | | Authors: | Goel, M, Kumari, S, Pal, R, Gupta, R. | | Deposit date: | 2017-08-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine
To Be Published
|
|
6ID6
 
 | |
5XC1
 
 | | Crystal structure of the complex of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 with S-1,2-Propanediol | | Descriptor: | Beta-xylanase, MAGNESIUM ION, S-1,2-PROPANEDIOL, ... | | Authors: | Bansia, H, Mahanta, P, Ramakumar, S. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
5XC0
 
 | |
2Q3N
 
 | | Agglutinin from Abrus Precatorius (APA-I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 A chain, Agglutinin-1 B chain | | Authors: | Bagaria, A, Surendranath, K, Ramagopal, U.A, Ramakumar, S, Karande, A.A. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Function Analysis and Insights into the Reduced Toxicity of Abrus precatorius Agglutinin I in Relation to Abrin.
J.Biol.Chem., 281, 2006
|
|
3OIW
 
 | | H-RasG12V with allosteric switch in the "on" state | | Descriptor: | ACETATE ION, CALCIUM ION, GTPase HRas, ... | | Authors: | Kumar, S, Buhrman, G, Mattos, C. | | Deposit date: | 2010-08-20 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Allosteric Modulation of Ras-GTP Is Linked to Signal Transduction through RAF Kinase.
J.Biol.Chem., 286, 2011
|
|
