8CPK
 
 | |
4I84
 
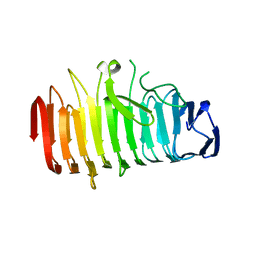 | | The crystal structure of the Haemophilus influenzae HxuA secretion domain involved in the two-partner secretion pathway | | Descriptor: | Heme/hemopexin-binding protein | | Authors: | Baelen, S, Dewitte, F, Clantin, B, Villeret, V. | | Deposit date: | 2012-12-03 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the secretion domain of HxuA from Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2ODL
 
 | |
1RWR
 
 | |
4RM6
 
 | | Crystal structure of Hemopexin Binding Protein | | Descriptor: | Heme/hemopexin-binding protein | | Authors: | Zambolin, S, Clantin, B, Haouz, A, Villeret, V, Delepelaire, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for haem piracy from host haemopexin by Haemophilus influenzae.
Nat Commun, 7, 2016
|
|
8VBB
 
 | |
4RT6
 
 | | Structure of a complex between hemopexin and hemopexin binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heme/hemopexin-binding protein, Hemopexin | | Authors: | Zambolin, S, Clantin, B, Haouz, A, Villeret, V, Delepelaire, P. | | Deposit date: | 2014-11-13 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for haem piracy from host haemopexin by Haemophilus influenzae.
Nat Commun, 7, 2016
|
|
