6BDJ
 
 | | Crystal structure of dioxygenase Tetur07g02040 | | Descriptor: | FE (III) ION, Tetur07g02040 | | Authors: | Daneshian, L, Schlachter, C.R, Chruszcz, M. | | Deposit date: | 2017-10-23 | | Release date: | 2018-11-14 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional characterization of an intradiol ring-cleavage dioxygenase from the polyphagous spider mite herbivore Tetranychus urticae Koch.
Insect Biochem.Mol.Biol., 107, 2019
|
|
4ILT
 
 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | CHLORIDE ION, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2012-12-31 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
5VG2
 
 | | Intradiol ring-cleavage Dioxygenase from Tetranychus urticae | | Descriptor: | CACODYLATE ION, FE (III) ION, Intradiol ring-cleavage Dioxygenase | | Authors: | Schlachter, C, Klapper, V, Chruszcz, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and functional characterization of an intradiol ring-cleavage dioxygenase from the polyphagous spider mite herbivore Tetranychus urticae Koch.
Insect Biochem.Mol.Biol., 107, 2019
|
|
2XSU
 
 | | Crystal structure of the A72G mutant of Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
2PCD
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE FROM PSEUDOMONAS AERUGINOSA AT 2.15 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE (ALPHA CHAIN), PROTOCATECHUATE 3,4-DIOXYGENASE (BETA CHAIN) | | Authors: | Ohlendorf, D.H, Orville, A.M, Lipscomb, J.D. | | Deposit date: | 1994-06-21 | | Release date: | 1994-12-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa at 2.15 A resolution.
J.Mol.Biol., 244, 1994
|
|
1TMX
 
 | | Crystal structure of hydroxyquinol 1,2-dioxygenase from Nocardioides Simplex 3E | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Travkin, V.M, Seifert, J, Schlomann, M, Golovleva, L, Scozzafava, A, Briganti, F. | | Deposit date: | 2004-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation.
J.Biol.Chem., 280, 2005
|
|
2XSR
 
 | | Crystal structure of wild type Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
2XSV
 
 | | Crystal structure of L69A mutant Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
1S9A
 
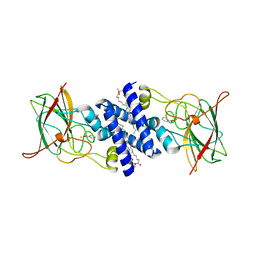 | | Crystal Structure of 4-Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP | | Descriptor: | (1-HEXADECANOYL-2-TETRADECANOYL-GLYCEROL-3-YL) PHOSPHONYL CHOLINE, BENZOIC ACID, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2004-02-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
J.Biol.Chem., 279, 2004
|
|
4ILV
 
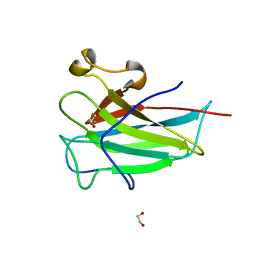 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
3T67
 
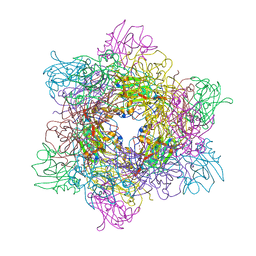 | |
3T63
 
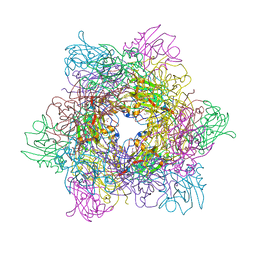 | |
3TH1
 
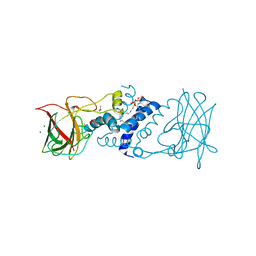 | | Crystal structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ACETATE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Rustiguel, J.K, Nonato, M.C. | | Deposit date: | 2011-08-18 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida
To be Published
|
|
5VXT
 
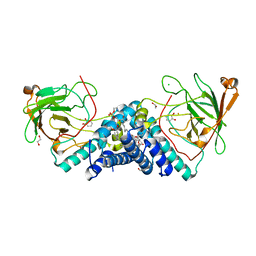 | |
5UMH
 
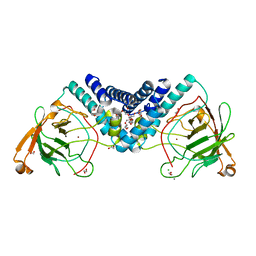 | |
5TD3
 
 | |
4WHQ
 
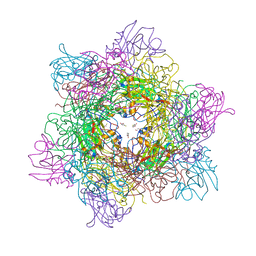 | | Alkylperoxo reaction intermediate trapped in Protocatechuate 3,4-dioxygenase (pseudomonas putida) at pH 6.5 | | Descriptor: | (6S)-4-fluoro-6-hydroperoxy-6-hydroxycyclohexa-2,4-dien-1-one, 4-fluorobenzene-1,2-diol, BETA-MERCAPTOETHANOL, ... | | Authors: | Knoot, C.J, Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WHO
 
 | |
4WHS
 
 | | 4-fluorocatechol bound to Protocatechuate 3,4-dioxygenase (pseudomonas putida) at pH 8.5 | | Descriptor: | 4-fluorobenzene-1,2-diol, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Knoot, C.J, Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WHP
 
 | |
4WHR
 
 | | Anhydride reaction intermediate trapped in Protocatechuate 3,4-dioxygenase (pseudomonas putida) at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-fluorobenzene-1,2-diol, 4-fluorooxepine-2,7-dione, ... | | Authors: | Knoot, C.J, Lipscomb, J.D. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3LMX
 
 | | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Purpero, V.M, Lipscomb, J.D, Shi, K. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published
|
|
3LKT
 
 | |
3LXV
 
 | | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-NITROCATECHOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2010-02-25 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published, 2010
|
|
1YKL
 
 | | Protocatechuate 3,4-Dioxygenase Y408C mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
