8TYG
 
 | | Plasmodium vivax PMV-WM08 inhibitor complex | | Descriptor: | (2E,4aR,7aS)-6-[3-(4-chlorophenyl)pyridin-2-yl]-7a-(2,5-difluorophenyl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hodder, A.N, Scally, S.W, Cowman, A.F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Plasmodium vivax PMV-WM08 inhibitor complex
To Be Published
|
|
8TYH
 
 | | Plasmodium vivax PMX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-tungstotellurate(VI), Aspartyl protease, ... | | Authors: | Hodder, A.N, Scally, S.W, Cowman, A.F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Plasmodium vivax PMX
To Be Published
|
|
8TYF
 
 | | Plasmodium vivax PMV-WM06 inhibitor complex | | Descriptor: | (2E,4aR,7aS)-6-[(3M)-3-(2-chlorophenyl)pyridin-2-yl]-7a-(2,5-difluorophenyl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hodder, A.N, Scally, S.W, Czabotar, P.E, Cowman, A.F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Plasmodium vivax PMX-XX inhibitor complex
To Be Published
|
|
8Q4B
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)-5-(3-(2-nitrophenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-163.1) | | Descriptor: | (3~{R},5~{R})-3-[2-[[ethynyl(methyl)amino]methyl]-1,3-thiazol-4-yl]-5-[3-(2-nitrophenyl)-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
7H57
 
 | | Crystal structure of endothiapepsin PF_cryo1 in complex with AC39729 at 100 K | | Descriptor: | 5-fluoranylpyridin-2-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5E
 
 | | Crystal structure of endothiapepsin PN_RT2 in complex with AC40075 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5B
 
 | | Crystal structure of endothiapepsin IS_RT2 in complex with AC39729 at 296 K | | Descriptor: | 5-fluoranylpyridin-2-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.122 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H58
 
 | | Crystal structure of endothiapepsin PF_RT2 in complex with AC39729 at 296 K | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H59
 
 | | Crystal structure of endothiapepsin IS_cryo3 in complex with AC39729 at 100 K | | Descriptor: | 5-fluoranylpyridin-2-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5C
 
 | | Crystal structure of endothiapepsin PN_cryo3 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5G
 
 | | Crystal structure of endothiapepsin PF_cryo1 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5H
 
 | | Crystal structure of endothiapepsin PF_RT2 in complex with AC40075 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5F
 
 | | Crystal structure of endothiapepsin PF_cryo3 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5K
 
 | | Crystal structure of endothiapepsin IS_RT2 in complex with AC40075 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.027 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5O
 
 | | Crystal structure of endothiapepsin PF_cryo3 in complex with TL00150 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5L
 
 | | Crystal structure of endothiapepsin PN_cryo3 in complex with TL00150 at 100 K | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, [4-(trifluoromethyl)phenyl]methanamine | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5R
 
 | | Crystal structure of endothiapepsin IS_cryo3 in complex with TL00150 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5M
 
 | | Crystal structure of endothiapepsin PN_cryo1 in complex with TL00150 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H56
 
 | | Crystal structure of endothiapepsin PF_cryo3 in complex with AC39729 at 100 K | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5P
 
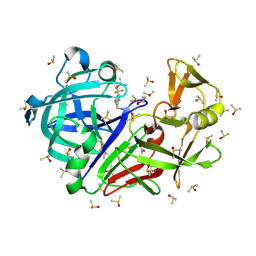 | | Crystal structure of endothiapepsin PF_cryo1 in complex with TL00150 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5I
 
 | | Crystal structure of endothiapepsin IS_cryo3 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5S
 
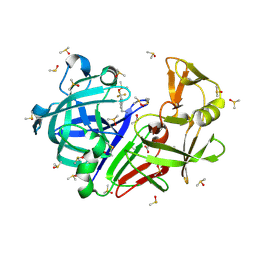 | | Crystal structure of endothiapepsin IS_cryo1 in complex with TL00150 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5A
 
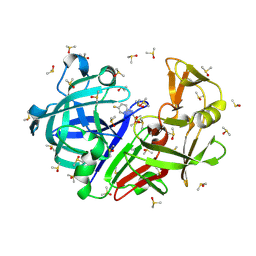 | | Crystal structure of endothiapepsin IS_cryo1 in complex with AC39729 at 100 K | | Descriptor: | 5-fluoranylpyridin-2-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5U
 
 | | Crystal structure of endothiapepsin PF_cryo3 in complex with JFD03909 at 100 K | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5D
 
 | | Crystal structure of endothiapepsin PN_cryo1 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
