9XIM
 
 | |
9XIA
 
 | |
9WGA
 
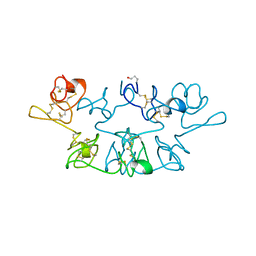 | |
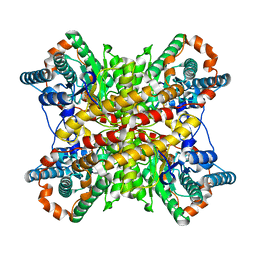
9RUB
 
 | | CRYSTAL STRUCTURE OF ACTIVATED RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE COMPLEXED WITH ITS SUBSTRATE, RIBULOSE-1,5-BISPHOSPHATE | | Descriptor: | FORMIC ACID, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, ... | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1990-11-28 | | Release date: | 1993-01-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate.
J.Biol.Chem., 266, 1991
|
|
9RSA
 
 | |
9RNT
 
 | |
9RAT
 
 | |
9PTI
 
 | |
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
9PAP
 
 | |
9PAI
 
 | | CLEAVED SUBSTRATE VARIANT OF PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PROTEIN (PLASMINOGEN ACTIVATOR INHIBITOR-1) residues 19-364, PROTEIN (PLASMINOGEN ACTIVATOR INHIBITOR-1) residues 365-397 | | Authors: | Aertgeerts, K, De Bondt, H.L, De Ranter, C.J, Declerck, P.J. | | Deposit date: | 1999-03-11 | | Release date: | 1999-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanisms contributing to the conformational and functional flexibility of plasminogen activator inhibitor-1.
Nat.Struct.Biol., 2, 1995
|
|
9NSE
 
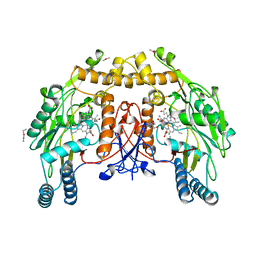 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, ETHYL-ISOSELENOUREA COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | Authors: | Li, H, Raman, C.S, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2000-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
9MSI
 
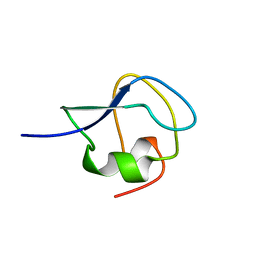 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18N | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
9MHT
 
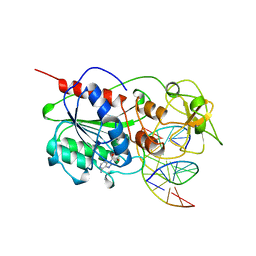 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-07 | | Release date: | 1998-12-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
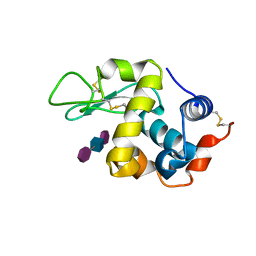
9LYZ
 
 | |
9LPR
 
 | |
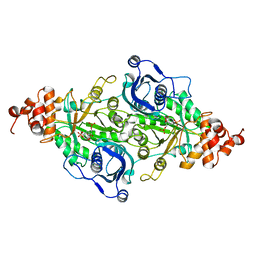
9LDT
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9LDB
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9JDW
 
 | |
9JDT
 
 | |
9JDQ
 
 | |
9JD2
 
 | |
9JBQ
 
 | | Structure of the complex between h1F3 Fab and PcrV fragment | | Descriptor: | Heavy chain, PcrV, light chain | | Authors: | Numata, S, Hara, T, Izawa, M, Okuno, Y, Sato, Y, Yamane, S, Maki, H, Sato, T, Yamano, Y. | | Deposit date: | 2024-08-27 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel humanized anti-PcrV monoclonal antibody, COT-143, protects mice from lethal Pseudomonas aeruginosa infection via inhibition of toxin translocation by the type III secretion system
To Be Published
|
|
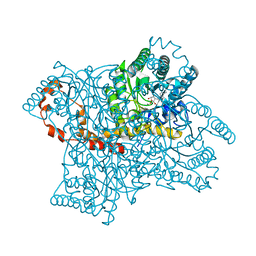
9JA1
 
 | | The RNA polymerase II elongation complex from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*CP*TP*CP*CP*TP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*T)-3'), DNA (5'-D(P*TP*GP*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*C)-3'), ... | | Authors: | Yi, G, Ma, J, Zhang, P. | | Deposit date: | 2024-08-23 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The RNA polymerase II elongation complex from Saccharomyces cerevisiae
To Be Published
|
|
9JA0
 
 | |
