2UYF
 
 | | Single mutant F111L DntR from Burkholderia sp. strain DNT in complex with thiocyanate | | Descriptor: | ACETATE ION, GLYCEROL, REGULATORY PROTEIN, ... | | Authors: | Lonneborg, R, Smirova, I, Dian, C, Leonard, G.A, Mcsweeney, S, Brzezinski, P. | | Deposit date: | 2007-04-04 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Vivo and in Vitro Investigation of Transcriptional Regulation by Dntr.
J.Mol.Biol., 372, 2007
|
|
2UYH
 
 | |
2UZ4
 
 | |
2V4M
 
 | | The isomerase domain of human glutamine-fructose-6-phosphate transaminase 1 (GFPT1) in complex with fructose 6-phosphate | | Descriptor: | CHLORIDE ION, FRUCTOSE -6-PHOSPHATE, GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE [ISOMERIZING] 1 | | Authors: | Moche, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, S, Schueler, H, Thorsell, A.G, Tresaugues, L, Uppenberg, J, Van Den Berg, S, Welin, M, Wisniewska, M, Weigelt, J, Nordlund, P, Wikstrom, M. | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Isomerase Domain of Human Gfpt1 in Complex with Fructose 6-Phosphate
To be Published
|
|
2VAD
 
 | | Monomeric red fluorescent protein, DsRed.M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RED FLUORESCENT PROTEIN, ... | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2VAE
 
 | | Fast maturing red fluorescent protein, DsRed.T4 | | Descriptor: | 1,2-ETHANEDIOL, RED FLUORESCENT PROTEIN | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2VMA
 
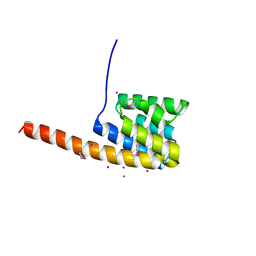 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F, IODIDE ION | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
2VMB
 
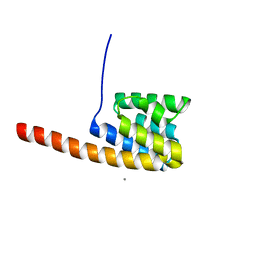 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
2VVH
 
 | | IrisFP fluorescent protein in its green form, cis conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VVI
 
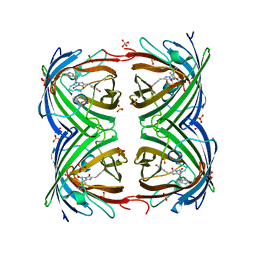 | | IrisFP fluorescent protein in its green form, trans conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VVJ
 
 | | IrisFP fluorescent protein in its red form, cis conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VZX
 
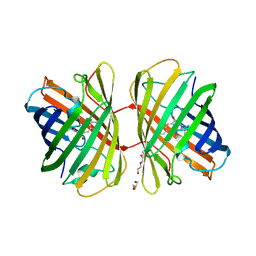 | | Structural and spectroscopic characterization of photoconverting fluorescent protein Dendra2 | | Descriptor: | GLYCEROL, Green fluorescent protein, TETRAETHYLENE GLYCOL | | Authors: | Adam, V, Nienhaus, K, Bourgeois, D, Nienhaus, G.U. | | Deposit date: | 2008-08-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Enhanced Photoconversion Yield in Green Fluorescent Protein-Like Protein Dendra2.
Biochemistry, 48, 2009
|
|
2W9Y
 
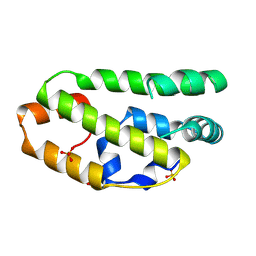 | | The structure of the lipid binding protein Ce-FAR-7 from Caenorhabditis elegans | | Descriptor: | FATTY ACID/RETINOL BINDING PROTEIN PROTEIN 7, ISOFORM A, CONFIRMED BY TRANSCRIPT EVIDENCE, ... | | Authors: | Jordanova, R, Groves, M.R, Tucker, P.A. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-20 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fatty Acid and Retinoid Binding Proteins Have Distinct Binding Pockets for the Two Types of Cargo
J.Biol.Chem., 284, 2009
|
|
2WSN
 
 | | Structure of Enhanced Cyan Fluorescent Protein at physiological pH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Lelimousin, M, Noirclerc-Savoye, M, Lazareno-Saez, C, Paetzold, B, Le Vot, S, Chazal, R, Macheboeuf, P, Field, M.J, Bourgeois, D, Royant, A. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Intrinsic Dynamics in Ecfp and Cerulean Control Fluorescence Quantum Yield.
Biochemistry, 48, 2009
|
|
2WSO
 
 | | Structure of Cerulean Fluorescent Protein at physiological pH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Lelimousin, M, Noirclerc-Savoye, M, Lazareno-Saez, C, Paetzold, B, Le Vot, S, Chazal, R, Macheboeuf, P, Field, M.J, Bourgeois, D, Royant, A. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Intrinsic Dynamics in Ecfp and Cerulean Control Fluorescence Quantum Yield.
Biochemistry, 48, 2009
|
|
2WUR
 
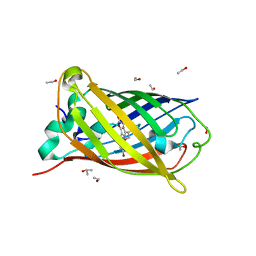 | |
2X26
 
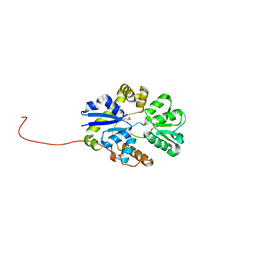 | | Crystal structure of the periplasmic aliphatic sulphonate binding protein SsuA from Escherichia coli | | Descriptor: | GLYCEROL, PERIPLASMIC ALIPHATIC SULPHONATES-BINDING PROTEIN | | Authors: | Beale, J, Lee, S, Iwata, S, Beis, K. | | Deposit date: | 2010-01-11 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Aliphatic Sulfonate-Binding Protein Ssua from Escherichia Coli
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2X9N
 
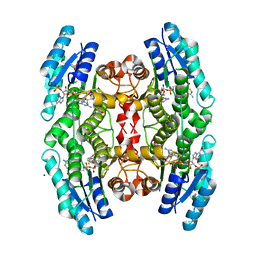 | | High resolution structure of TbPTR1 in complex with cyromazine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, ACETATE ION, DITHIANE DIOL, ... | | Authors: | Dawson, A, Tulloch, L.B, Barrack, K.L, Hunter, W.N. | | Deposit date: | 2010-03-23 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Resolution Structures of Trypanosoma Brucei Pteridine Reductase Ligand Complexes Inform on the Placement of New Molecular Entities in the Active Site of a Potential Drug Target
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XB4
 
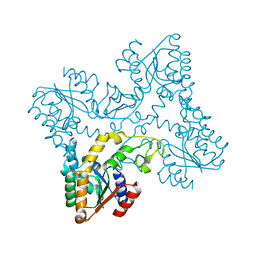 | | Crystal structures of zinc containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | ADENYLATE KINASE, S,R MESO-TARTARIC ACID, ZINC ION | | Authors: | Mukhopadhyay, A, Kladova, A.V, Gavel, O.Y, Calvete, J.J, Shnyrov, V.L, Moura, I, Moura, J.J.G, Bursakov, S.A, Romao, M.J, Trincao, J. | | Deposit date: | 2010-04-05 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Zinc-, Cobalt-, and Iron-Containing Adenylate Kinase from Desulfovibrio Gigas: A Novel Metal-Containing Adenylate Kinase from Gram-Negative Bacteria.
J.Biol.Inorg.Chem., 16, 2011
|
|
2XUT
 
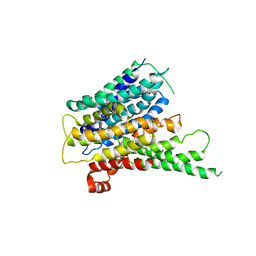 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
2XVY
 
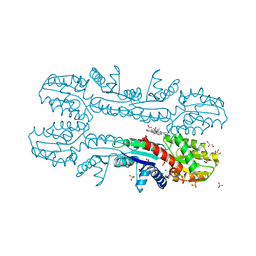 | | Cobalt chelatase CbiK (periplasmic) from Desulvobrio vulgaris Hildenborough (co-crystallised with cobalt and SHC) | | Descriptor: | CHELATASE, PUTATIVE, COBALT (II) ION, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Desulfovibrio vulgaris CbiK(P) cobaltochelatase: evolution of a haem binding protein orchestrated by the incorporation of two histidine residues.
Environ. Microbiol., 19, 2017
|
|
2Y0G
 
 | |
2YDZ
 
 | | X-ray structure of the cyan fluorescent protein SCFP3A (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
2YE1
 
 | | X-ray structure of the cyan fluorescent proteinmTurquoise-GL (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of the Cyan Fluorescent Protein Mturquoise-Gl
To be Published
|
|
