1LRP
 
 | |
1R69
 
 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | REPRESSOR PROTEIN CI | | 著者 | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | 登録日 | 1988-12-08 | | 公開日 | 1989-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
2CRO
 
 | |
2OR1
 
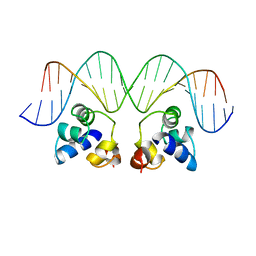 | | RECOGNITION OF A DNA OPERATOR BY THE REPRESSOR OF PHAGE 434. A VIEW AT HIGH RESOLUTION | | 分子名称: | 434 REPRESSOR, DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3') | | 著者 | Aggarwal, A.K, Rodgers, D.W, Drottar, M, Ptashne, M, Harrison, S.C. | | 登録日 | 1989-09-05 | | 公開日 | 1989-09-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Recognition of a DNA operator by the repressor of phage 434: a view at high resolution.
Science, 242, 1988
|
|
3CRO
 
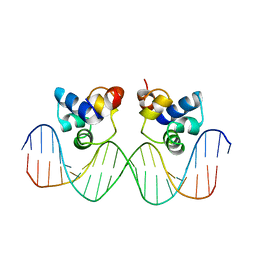 | |
1LMB
 
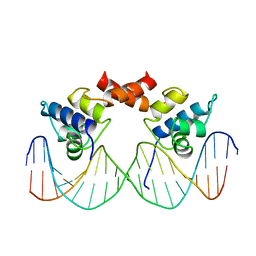 | |
1PRA
 
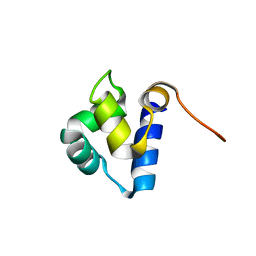 | |
1RPE
 
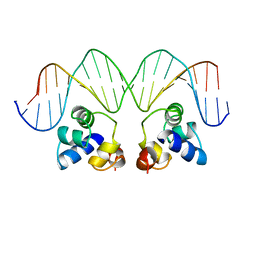 | |
1ADR
 
 | |
1PER
 
 | |
1LLI
 
 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | 分子名称: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | 著者 | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | 登録日 | 1994-03-25 | | 公開日 | 1994-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-08 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
2R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-13 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1ZUG
 
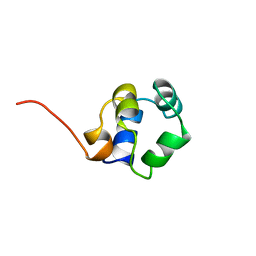 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | 分子名称: | PHAGE 434 CRO PROTEIN | | 著者 | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | 登録日 | 1997-03-14 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
1B0N
 
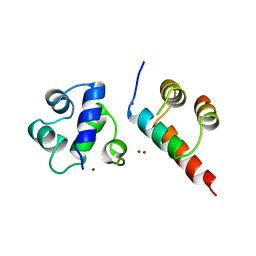 | | SINR PROTEIN/SINI PROTEIN COMPLEX | | 分子名称: | PROTEIN (SINI PROTEIN), PROTEIN (SINR PROTEIN), ZINC ION | | 著者 | Lewis, R.J, Brannigan, J.A, Offen, W.A, Smith, I, Wilkinson, A.J. | | 登録日 | 1998-11-11 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex.
J.Mol.Biol., 283, 1998
|
|
1RIO
 
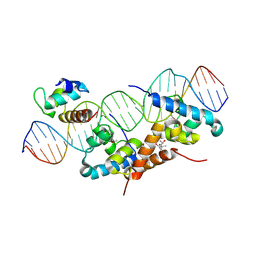 | | Structure of bacteriophage lambda cI-NTD in complex with sigma-region4 of Thermus aquaticus bound to DNA | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 27-MER, CALCIUM ION, ... | | 著者 | Jain, D, Nickels, B.E, Sun, L, Hochschild, A, Darst, S.A. | | 登録日 | 2003-11-17 | | 公開日 | 2004-01-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of a ternary transcription activation complex.
Mol.Cell, 13, 2004
|
|
1UTX
 
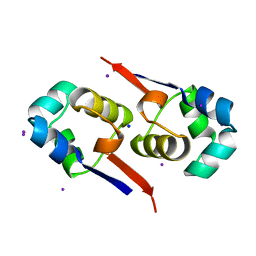 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | 分子名称: | CYLR2, IODIDE ION, SODIUM ION | | 著者 | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | 登録日 | 2003-12-12 | | 公開日 | 2004-09-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
1SQ8
 
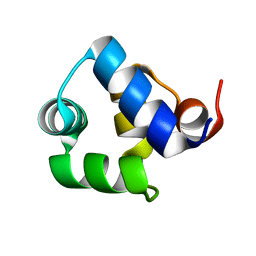 | |
1Y7Y
 
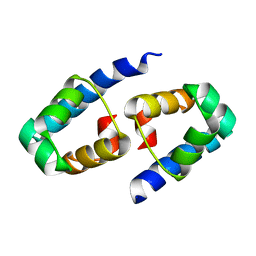 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | 分子名称: | C.AhdI | | 著者 | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | 登録日 | 2004-12-10 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
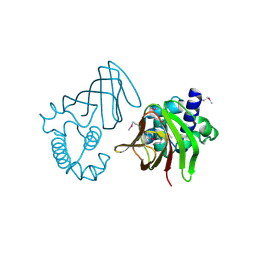
1Y9Q
 
 | |
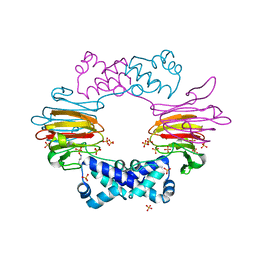
2BNM
 
 | |
2BNO
 
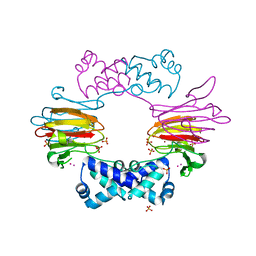 | | The structure of Hydroxypropylphosphonic acid epoxidase from S. wedmorenis. | | 分子名称: | EPOXIDASE, MERCURY (II) ION, SULFATE ION, ... | | 著者 | McLuskey, K, Cameron, S, Hunter, W.N. | | 登録日 | 2005-03-29 | | 公開日 | 2005-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and Reactivity of Hydroxypropylphosphonic Acid Epoxidase in Fosfomycin Biosynthesis by a Cation- and Flavin-Dependent Mechanism.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BNN
 
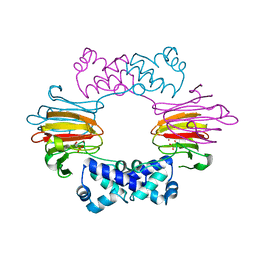 | |
1X57
 
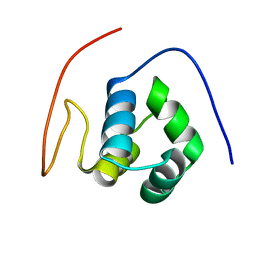 | | Solution structures of the HTH domain of human EDF-1 protein | | 分子名称: | Endothelial differentiation-related factor 1 | | 著者 | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-15 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of the HTH domain of human EDF-1 protein
To be Published
|
|
1ZZ6
 
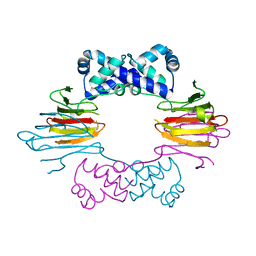 | | Crystal Structure of Apo-HppE | | 分子名称: | Hydroxypropylphosphonic Acid Epoxidase | | 著者 | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | 登録日 | 2005-06-13 | | 公開日 | 2005-07-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
