8YWQ
 
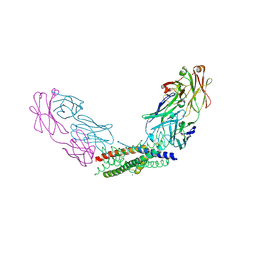 | | Crystal structure of the Fab fragment of the anti-IL-6 antibody I9H in complex with a domain-swapped IL-6 dimer | | Descriptor: | Heavy chain of the Fab fragment of anti-IL-6 antibody I9H, Interleukin-6, Light chain of the Fab fragment of anti-IL-6 antibody I9H, ... | | Authors: | Bukhdruker, S, Yudenko, A, Marin, E, Remeeva, A, Rodin, S, Burtseva, A, Petrov, A, Ischenko, A, Borshchevskiy, V. | | Deposit date: | 2024-03-31 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of signaling complex inhibition by IL-6 domain-swapped dimers
Structure, 2025
|
|
8YWR
 
 | |
8Z2G
 
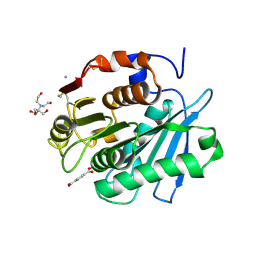 | | MHET bound form of PET-degrading cutinase mutant Cut190*SS_S176A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, AMMONIUM ION, ... | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2H
 
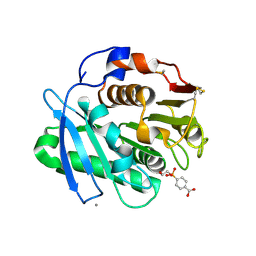 | | Substrate analog a010 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 4-[2-hydroxyethyloxy(oxidanyl)phosphoryl]benzoic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2I
 
 | | Substrate analog a011 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 2-hydroxyethyloxy-(4-methoxycarbonylphenyl)phosphinic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2J
 
 | | Substrate analog a012 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | (4-methoxycarbonylphenyl)-(2-phenylmethoxyethoxy)phosphinic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2K
 
 | | Substrate analog a013 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 4-[oxidanyl(2-phenylmethoxyethoxy)phosphoryl]benzoic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2W
 
 | | Crystal structure of apo- exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) | | Descriptor: | 2-methylpropylphosphonic acid, Glucan 1,3-beta-glucosidase A, SODIUM ION | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 2024
|
|
8Z2X
 
 | | Crystal structure of exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) as a complex with cellobiose | | Descriptor: | Glucan 1,3-beta-glucosidase A, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 2024
|
|
8Z2Y
 
 | | High-resolution crystal structure of exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) as a complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 2024
|
|
8ZNS
 
 | |
8ZUP
 
 | |
8ZUQ
 
 | |
8ZUR
 
 | |
8ZUS
 
 | |
8ZUT
 
 | |
8ZX0
 
 | | Crystal Structure of CntL in complex with a dual-site inhibitor | | Descriptor: | (2S)-2-[2-[[(2S,3S,4R,5R)-5-(6-azanyl-2-chloranyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]ethylamino]-3-(1H-imidazol-4-yl)propanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Luo, Z, Zhou, H. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structure-guided inhibitor design targeting CntL provides the first chemical validation of the staphylopine metallophore system in bacterial metal acquisition.
Eur.J.Med.Chem., 280, 2024
|
|
9AUQ
 
 | | Crystal structure of 4-Fluoro-tryptophan labeled Oscillatoria Agardhii agglutinin | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Lectin | | Authors: | Runge, B.R, Zadorozhnyi, R.R, Quinn, C.M, Russell, R.W, Lu, M, Antolinez, S, Struppe, J, Schwieters, C.D, Byeon, I.L, Hadden-Perilla, J, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2024-02-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Integrating 19 F Distance Restraints for Accurate Protein Structure Determination by Magic Angle Spinning NMR Spectroscopy.
J.Am.Chem.Soc., 2024
|
|
9AYT
 
 | | Structure of the quorum quenching lactonase GcL bound to N-hexanoyl-L-homoserine lactone | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
9B2I
 
 | | Structure of the quorum quenching lactonase GcL G156P mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
9B2J
 
 | | Structure of the quorum quenching lactonase GcL I237M mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
9B2L
 
 | | Structure of the quorum quenching lactonase GcL D122N mutant - bimetallic center | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
9B2N
 
 | | Structure of the quorum quenching lactonase GcL D122N mutant - monometal center | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
9B2O
 
 | | Structure of the quorum quenching lactonase GcL bound to the hydrolysis product of N-octanoyl-L-homoserine lactone | | Descriptor: | COBALT (II) ION, FE (III) ION, GcL lactonase, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
9B2P
 
 | | Structure of the quorum quenching lactonase GcL D122N mutant - bimetallic metal center - C2 space group | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (III) ION, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
