1QT6
 
 | | E11H Mutant of T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT7
 
 | | E11N Mutant of T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT8
 
 | | T26H Mutant of T4 Lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT9
 
 | | OXIDIZED [2FE-2S] FERREDOXIN FROM ANABAENA PCC7119 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Morales, R, Chron, M.-H, Hudry-Clergeon, G, Petillot, Y, Norager, S, Medina, M, Frey, M. | | Deposit date: | 1999-07-01 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refined X-ray structures of the oxidized, at 1.3 A, and reduced, at 1.17 A, [2Fe-2S] ferredoxin from the cyanobacterium Anabaena PCC7119 show redox-linked conformational changes.
Biochemistry, 38, 1999
|
|
1QTB
 
 | |
1QTC
 
 | |
1QTD
 
 | |
1QTE
 
 | | CRYSTAL STRUCTURE OF THE 70 KDA SOLUBLE LYTIC TRANSGLYCOSYLASE SLT70 FROM ESCHERICHIA COLI AT 1.90 A RESOLUTION IN COMPLEX WITH A 1,6-ANHYDROMUROTRIPEPTIDE | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | van Asselt, E.J, Thunnissen, A.-M.W.H, Dijkstra, B.W. | | Deposit date: | 1999-06-27 | | Release date: | 1999-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment.
J.Mol.Biol., 291, 1999
|
|
1QTH
 
 | |
1QTI
 
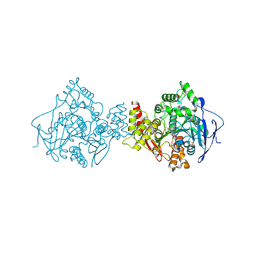 | | Acetylcholinesterase (E.C.3.1.1.7) | | Descriptor: | (-)-GALANTHAMINE, ACETYLCHOLINESTERASE | | Authors: | Bartolucci, C, Perola, E, Pilger, C, Fels, G, Lamba, D. | | Deposit date: | 1999-06-28 | | Release date: | 1999-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of a complex of galanthamine (Nivalin) with acetylcholinesterase from Torpedo californica: implications for the design of new anti-Alzheimer drugs
Proteins, 42, 2001
|
|
1QTQ
 
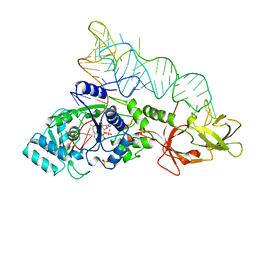 | | GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH TRNA AND AN AMINO ACID ANALOG | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, PROTEIN (GLUTAMINYL-TRNA SYNTHETASE), RNA (TRNA GLN II ), ... | | Authors: | Rath, V.L, Silvian, L.F, Beijer, B, Sproat, B.S, Steitz, T.A. | | Deposit date: | 1998-01-28 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How glutaminyl-tRNA synthetase selects glutamine.
Structure, 6, 1998
|
|
1QTV
 
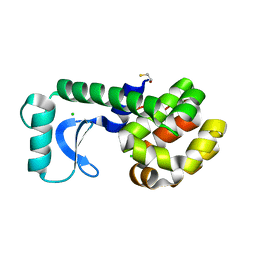 | | T26E APO STRUCTURE OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QTX
 
 | |
1QTZ
 
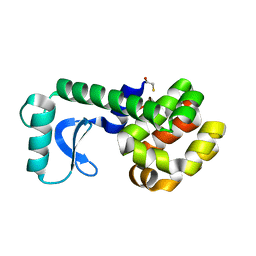 | | D20C MUTANT OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QU0
 
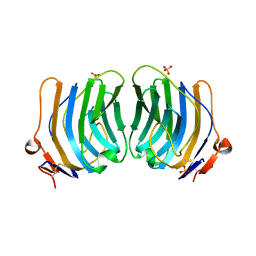 | | CRYSTAL STRUCTURE OF THE FIFTH LAMININ G-LIKE MODULE OF THE MOUSE LAMININ ALPHA2 CHAIN | | Descriptor: | CALCIUM ION, LAMININ ALPHA2 CHAIN, SULFATE ION | | Authors: | Hohenester, E, Tisi, D, Talts, J.F, Timpl, R. | | Deposit date: | 1999-07-05 | | Release date: | 1999-12-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin.
Mol.Cell, 4, 1999
|
|
1QU1
 
 | | CRYSTAL STRUCTURE OF EHA2 (23-185) | | Descriptor: | PROTEIN (INFLUENZA RECOMBINANT HA2 CHAIN) | | Authors: | Chen, J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-07-05 | | Release date: | 2000-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QU2
 
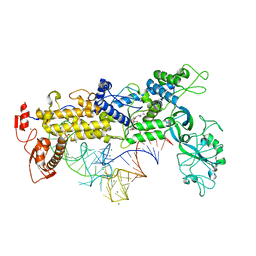 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
1QU3
 
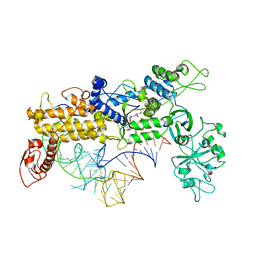 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MUPIROCIN, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
1QU7
 
 | |
1QU9
 
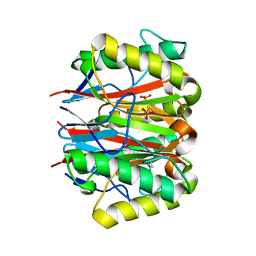 | | 1.2 A CRYSTAL STRUCTURE OF YJGF GENE PRODUCT FROM E. COLI | | Descriptor: | YJGF PROTEIN | | Authors: | Volz, K. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-01 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A test case for structure-based functional assignment: the 1.2 A crystal structure of the yjgF gene product from Escherichia coli
Protein Sci., 8, 1999
|
|
1QUB
 
 | | CRYSTAL STRUCTURE OF THE GLYCOSYLATED FIVE-DOMAIN HUMAN BETA2-GLYCOPROTEIN I PURIFIED FROM BLOOD PLASMA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (human beta2-Glycoprotein I), ... | | Authors: | Bouma, B, de Groot, P.G, van den Elsen, J.M.H, Ravelli, R.B.G, Schouten, A, Simmelink, M.J.A, Derksen, R.H.W.M, Kroon, J, Gros, P. | | Deposit date: | 1999-07-01 | | Release date: | 1999-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure.
EMBO J., 18, 1999
|
|
1QUD
 
 | | L99G MUTANT OF T4 LYSOZYME | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, PROTEIN (LYSOZYME) | | Authors: | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | Deposit date: | 1999-07-01 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
1QUG
 
 | | E108V MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | Deposit date: | 1999-07-01 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
1QUH
 
 | | L99G/E108V MUTANT OF T4 LYSOZYME | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, PROTEIN (LYSOZYME) | | Authors: | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | Deposit date: | 1999-07-01 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
1QUI
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH BROMINE AND PHOSPHATE | | Descriptor: | BROMIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
