1E3U
 
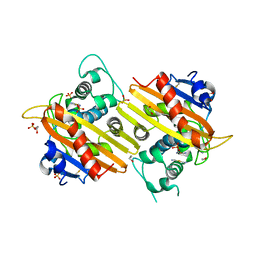 | | MAD structure of OXA10 class D beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-06-23 | | Release date: | 2001-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
1E3X
 
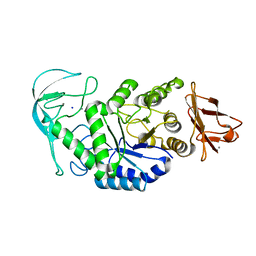 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.92A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-26 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E3Z
 
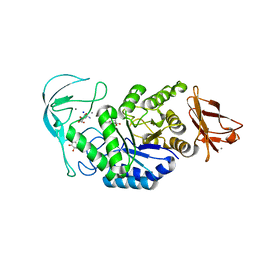 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E40
 
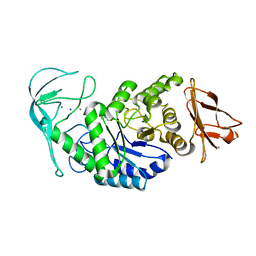 | | Tris/maltotriose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 2.2A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E42
 
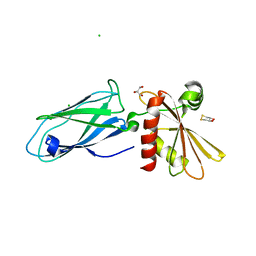 | | Beta2-adaptin appendage domain, from clathrin adaptor AP2 | | Descriptor: | AP-2 COMPLEX SUBUNIT BETA, CHLORIDE ION, DITHIANE DIOL, ... | | Authors: | Owen, D.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-06-27 | | Release date: | 2000-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure and Function of the Beta2-Adaptin Appendage Domain
Embo J., 19, 2000
|
|
1E43
 
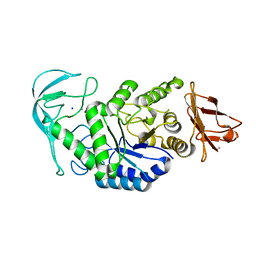 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E44
 
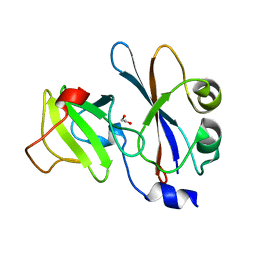 | | ribonuclease domain of colicin E3 in complex with its immunity protein | | Descriptor: | 1,2-ETHANEDIOL, COLICIN E3, IMMUNITY PROTEIN | | Authors: | Carr, S, Walker, D, James, R, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of a Ribosome Inactivating Ribonuclease: The Crystal Structure of the Cytotoxic Domain of Colicin E3 in Complex with its Immunity Protein
Structure, 8, 2000
|
|
1E46
 
 | |
1E47
 
 | |
1E48
 
 | |
1E49
 
 | |
1E4A
 
 | |
1E4B
 
 | |
1E4C
 
 | |
1E4D
 
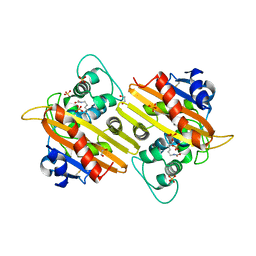 | | Structure of OXA10 beta-lactamase at pH 8.3 | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, SULFATE ION | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-07-03 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
1E4E
 
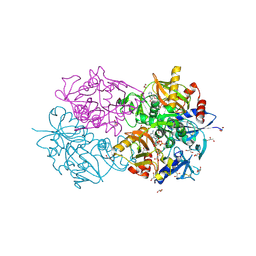 | | D-alanyl-D-lacate ligase | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roper, D.I. | | Deposit date: | 2000-07-03 | | Release date: | 2001-06-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular basis of vancomycin resistance in clinically relevant Enterococci: crystal structure of D-alanyl-D-lactate ligase (VanA).
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
1E4F
 
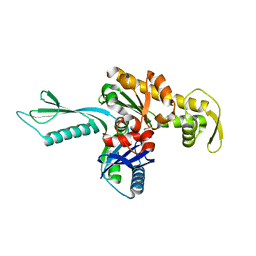 | | FtsA (apo form) from Thermotoga maritima | | Descriptor: | CELL DIVISION PROTEIN FTSA | | Authors: | van den Ent, F, Lowe, J. | | Deposit date: | 2000-07-03 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Cell Division Protein Ftsa from Thermotoga Maritima
Embo J., 19, 2000
|
|
1E4G
 
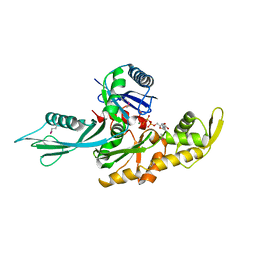 | | FtsA (ATP-bound form) from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN FTSA, MAGNESIUM ION | | Authors: | van den Ent, F, Lowe, J. | | Deposit date: | 2000-07-03 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Cell Division Protein Ftsa from Thermotoga Maritima
Embo J., 19, 2000
|
|
1E4L
 
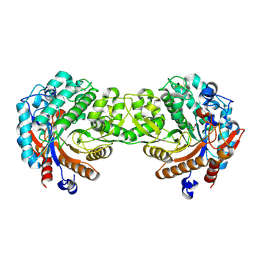 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZM Glu191Asp | | Descriptor: | BETA-GLUCOSIDASE, CHLOROPLASTIC, GLYCEROL | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-10 | | Release date: | 2000-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E4M
 
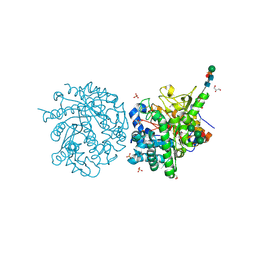 | | MYROSINASE FROM SINAPIS ALBA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-07-10 | | Release date: | 2001-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High Resolution X-Ray Crystallography Shows that Ascorbate is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base
J.Biol.Chem., 275, 2000
|
|
1E4N
 
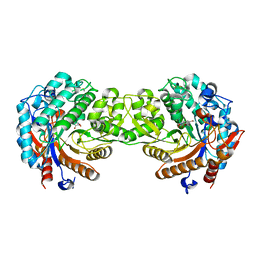 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-11 | | Release date: | 2000-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E4W
 
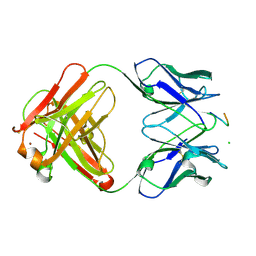 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CHLORIDE ION, CYCLIC PEPTIDE, NICKEL (II) ION, ... | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
1E4X
 
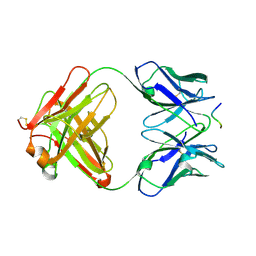 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CYCLIC PEPTIDE, TAB2 | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
1E4Y
 
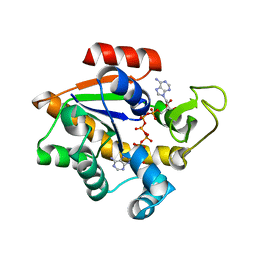 | |
1E50
 
 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
