1LTR
 
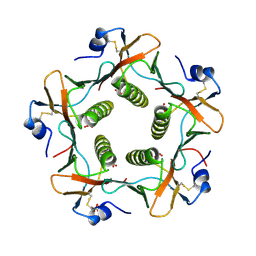 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HUMAN HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SULFATE ION | | Authors: | Matkovic-Calogovic, D, Loreggian, A, Palu, G, Zanotti, G. | | Deposit date: | 1998-07-31 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
1LTS
 
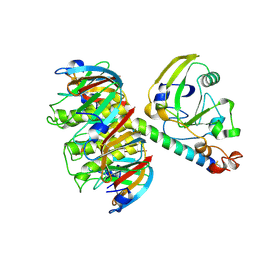 | |
1LTU
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTV
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE WITH BOUND OXIDIZED Fe(III) | | Descriptor: | FE (III) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTW
 
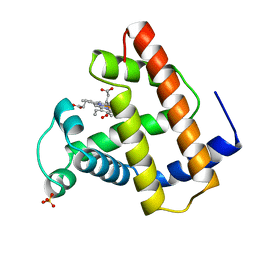 | | RECOMBINANT SPERM WHALE MYOGLOBIN 29W MUTANT (OXY) | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, T, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1996-11-02 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Perturbation of the Fe-O2 Bond by Nearby Residues in Heme Pocket: Observation of Vfe-O2 Raman Bands for Oxymyoglobin Mutants
J.Am.Chem.Soc., 118, 1996
|
|
1LTX
 
 | | Structure of Rab Escort Protein-1 in complex with Rab geranylgeranyl transferase and isoprenoid | | Descriptor: | AAAA, CHLORIDE ION, FARNESYL, ... | | Authors: | Pylypenko, O, Rak, A, Reents, R, Niculae, A, Thoma, N.H, Waldmann, H, Schlichting, I, Goody, R.S, Alexandrov, K. | | Deposit date: | 2002-05-21 | | Release date: | 2003-05-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase
Mol.Cell, 11, 2003
|
|
1LTZ
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LU0
 
 | | Atomic Resolution Structure of Squash Trypsin Inhibitor: Unexpected Metal Coordination | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Thaimattam, R, Tykarska, E, Bierzynski, A, Sheldrick, G.M, Jaskolski, M. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution structure of squash trypsin inhibitor: unexpected metal coordination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LU1
 
 | | THE STRUCTURE OF THE DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH THE FORSSMAN DISACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ADENINE, CALCIUM ION, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Strecker, G, Imberty, A, Fernandez, E, Wyns, L, Etzler, M.E. | | Deposit date: | 1998-07-24 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1LU2
 
 | | DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH THE BLOOD GROUP A TRISACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Strecker, G, Imberty, A, Fernandez, E, Wyns, L, Etzler, M.E. | | Deposit date: | 1998-07-30 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1LU4
 
 | | 1.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED MYCOBACTERIUM TUBERCULOSIS DISULFIDE OXIDOREDUCTASE HOMOLOGOUS TO E. COLI DSBE: IMPLICATIONS FOR FUNCTIONS | | Descriptor: | SOLUBLE SECRETED ANTIGEN MPT53 | | Authors: | Goulding, C.W, Apostol, M.I, Gleiter, S, Parseghian, A, Bardwell, J, Gennaro, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-21 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Gram-positive DsbE Proteins Function Differently from Gram-negative DsbE Homologs: A STRUCTURE TO FUNCTION ANALYSIS OF DsbE FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 279, 2004
|
|
1LU9
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
1LUA
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 complexed with NADP | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
1LUE
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68A/D122N MUTANT (MET) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-05-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
1LUF
 
 | | Crystal Structure of the MuSK Tyrosine Kinase: Insights into Receptor Autoregulation | | Descriptor: | muscle-specific tyrosine kinase receptor musk | | Authors: | Till, J.H, Becerra, M, Watty, A, Lu, Y, Ma, Y, Neubert, T.A, Burden, S.J, Hubbard, S.R. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation.
Structure, 10, 2002
|
|
1LUL
 
 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1LUR
 
 | | Crystal Structure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66 | | Descriptor: | SULFATE ION, aldose 1-epimerase | | Authors: | Keller, J.P, Xiao, R, MacDonald, L, Shen, J, Acton, T, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-23 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66
TO BE PUBLISHED
|
|
1LV0
 
 | | Crystal structure of the Rab effector guanine nucleotide dissociation inhibitor (GDI) in complex with a geranylgeranyl (GG) peptide | | Descriptor: | GERAN-8-YL GERAN, RAB GDP disossociation inhibitor alpha, SULFATE ION | | Authors: | An, Y, Shao, Y, Alory, C, Matteson, J, Sakisaka, T, Chen, W, Gibbs, R.A, Wilson, I.A, Balch, W.E. | | Deposit date: | 2002-05-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Geranylgeranyl switching regulates GDI-Rab GTPase recycling.
Structure, 11, 2003
|
|
1LV1
 
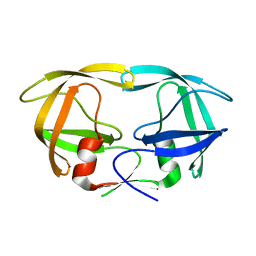 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | Descriptor: | HIV-1 protease | | Authors: | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1LV5
 
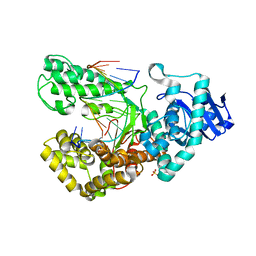 | | Crystal Structure of the Closed Conformation of Bacillus DNA Polymerase I Fragment Bound to DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*CP*GP*TP*CP*GP*CP*TP*GP*AP*TP*CP*CP*G)-3', 5'-D(*GP*GP*AP*TP*CP*AP*GP*CP*GP*A)-3', ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-05-24 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1LV7
 
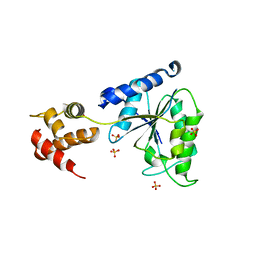 | | Crystal Structure of the AAA domain of FtsH | | Descriptor: | FtsH, SULFATE ION | | Authors: | Krzywda, S, Brzozowski, A.M, Verma, C, Karata, K, Ogura, T, Wilkinson, A.J. | | Deposit date: | 2002-05-26 | | Release date: | 2002-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution.
Structure, 10, 2002
|
|
1LVA
 
 | |
1LVB
 
 | | CATALYTICALLY INACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH SUBSTRATE | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), GLYCEROL, OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
1LVC
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 2' deoxy, 3' anthraniloyl ATP | | Descriptor: | 3'ANTHRANILOYL-2'-DEOXY-ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, YTTERBIUM (III) ION, ... | | Authors: | Shen, Y, Lee, Y.-S, Soelaiman, S, Bergson, P, Lu, D, Chen, A, Beckingham, K, Grabarek, Z, Mrksich, M, Tang, W.-J. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins
Embo J., 21, 2002
|
|
1LVE
 
 | |
