1DBG
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
1DBH
 
 | | DBL AND PLECKSTRIN HOMOLOGY DOMAINS FROM HSOS1 | | Descriptor: | PROTEIN (HUMAN SOS 1) | | Authors: | Soisson, S.M, Kuriyan, J. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein.
Cell(Cambridge,Mass.), 95, 1998
|
|
1DBI
 
 | | CRYSTAL STRUCTURE OF A THERMOSTABLE SERINE PROTEASE | | Descriptor: | AK.1 SERINE PROTEASE, CALCIUM ION, SODIUM ION | | Authors: | Smith, C.A, Toogood, H.S, Baker, H.M, Daniel, R.M, Baker, E.N. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calcium-mediated thermostability in the subtilisin superfamily: the crystal structure of Bacillus Ak.1 protease at 1.8 A resolution.
J.Mol.Biol., 294, 1999
|
|
1DBN
 
 | | MAACKIA AMURENSIS LEUKOAGGLUTININ (LECTIN) WITH SIALYLLACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Imberty, A, Gautier, C, Lescar, J, Loris, R. | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | An unusual carbohydrate binding site revealed by the structures of two Maackia amurensis lectins complexed with sialic acid-containing oligosaccharides.
J.Biol.Chem., 275, 2000
|
|
1DBO
 
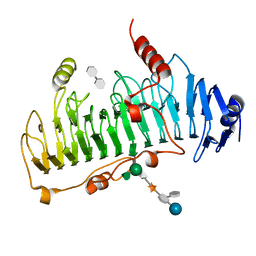 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-03 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
1DBP
 
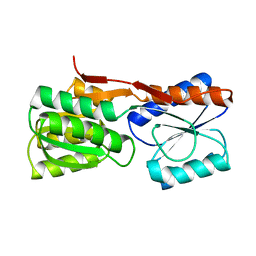 | |
1DBV
 
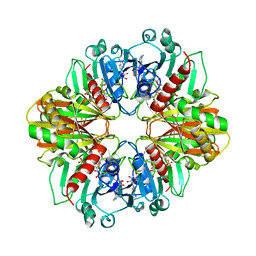 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1DBW
 
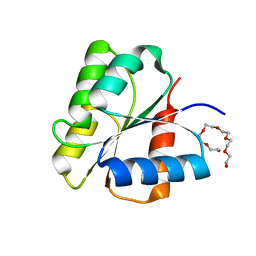 | | CRYSTAL STRUCTURE OF FIXJ-N | | Descriptor: | POLYETHYLENE GLYCOL (N=34), TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1DC0
 
 | |
1DC1
 
 | | RESTRICTION ENZYME BSOBI/DNA COMPLEX STRUCTURE: ENCIRCLEMENT OF THE DNA AND HISTIDINE-CATALYZED HYDROLYSIS WITHIN A CANONICAL RESTRICTION ENZYME FOLD | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, BSOBI RESTRICTION ENDONUCLEASE, DNA (5'-D(*T*AP*TP*AP*CP*TP*CP*GP*AP*GP*TP*AP*T)-3') | | Authors: | van der Woerd, M.J, Pelletier, J.J, Xu, S.-Y, Friedman, A.M. | | Deposit date: | 1999-11-04 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Restriction enzyme BsoBI-DNA complex: a tunnel for recognition of degenerate DNA sequences and potential histidine catalysis.
Structure, 9, 2001
|
|
1DC3
 
 | |
1DC4
 
 | | STRUCTURAL ANALYSIS OF GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE FROM ESCHERICHIA COLI: DIRECT EVIDENCE FOR SUBSTRATE BINDING AND COFACTOR-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Yun, M, Park, C.-G, Kim, J.-Y, Park, H.-W. | | Deposit date: | 1999-11-04 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes.
Biochemistry, 39, 2000
|
|
1DC5
 
 | |
1DC6
 
 | | STRUCTURAL ANALYSIS OF GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE FROM ESCHERICHIA COLI: DIRECT EVIDENCE FOR SUBSTRATE BINDING AND COFACTOR-INDUCED CONFORMATIONAL CHANGES. | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yun, M, Park, C.-G, Kim, J.-Y, Park, H.-W. | | Deposit date: | 1999-11-04 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes.
Biochemistry, 39, 2000
|
|
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
1DCF
 
 | |
1DCK
 
 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N COMPLEXED WITH MN2+ | | Descriptor: | MANGANESE (II) ION, POLYETHYLENE GLYCOL (N=34), TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1DCL
 
 | |
1DCM
 
 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N WITH AN ATYPICAL CONFORMER (MONOMER A) | | Descriptor: | MAGNESIUM ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1DCN
 
 | | INACTIVE MUTANT H162N OF DELTA 2 CRYSTALLIN WITH BOUND ARGININOSUCCINATE | | Descriptor: | ARGININOSUCCINATE, DELTA 2 CRYSTALLIN | | Authors: | Vallee, F, Turner, M.A, Lindley, P, Howell, P.L. | | Deposit date: | 1998-10-29 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an inactive duck delta II crystallin mutant with bound argininosuccinate.
Biochemistry, 38, 1999
|
|
1DCQ
 
 | | CRYSTAL STRUCTURE OF THE ARF-GAP DOMAIN AND ANKYRIN REPEATS OF PAPBETA. | | Descriptor: | PYK2-ASSOCIATED PROTEIN BETA, ZINC ION | | Authors: | Mandiyan, V, Andreev, J, Schlessinger, J, Hubbard, S.R. | | Deposit date: | 1999-11-05 | | Release date: | 1999-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein beta.
EMBO J., 18, 1999
|
|
1DCR
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1DCS
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE FROM S. CLAVULIGERUS | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, SULFATE ION | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
1DCU
 
 | | REDOX SIGNALING IN THE CHLOROPLAST: STRUCTURE OF OXIDIZED PEA FRUCTOSE-1,6-BISPHOSPHATE PHOSPHATASE | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Chiadmi, M, Navaza, A, Miginiac-Maslow, M, Jacquot, J.P, Cherfils, J. | | Deposit date: | 1999-11-05 | | Release date: | 1999-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
EMBO J., 18, 1999
|
|
1DCV
 
 | |
