1ODG
 
 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | Descriptor: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | Authors: | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
1ODI
 
 | |
1ODJ
 
 | |
1ODK
 
 | |
1ODL
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, GLYCEROL, PURINE NUCLEOSIDE PHOSPHORYLASE, ... | | Authors: | Tahirov, T.H, Inagaki, E, Miyano, M. | | Deposit date: | 2003-02-19 | | Release date: | 2003-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase from Thermus Thermophilus
J.Mol.Biol., 337, 2004
|
|
1ODM
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
1ODN
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN-EXPOSED PRODUCT FROM ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | 6-(5-AMINO-5-CARBOXY-PENTANOYLAMINO)-3-HYDROXYMETHYL-7-OXO-4-THIA-1-AZA-BICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
1ODO
 
 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
1ODS
 
 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1ODT
 
 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1ODU
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH FUCOSE | | Descriptor: | PUTATIVE ALPHA-L-FUCOSIDASE, beta-L-fucopyranose | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
1ODV
 
 | | Photoactive yellow protein 1-25 deletion mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Vreede, J, Van Der horst, M.A, Hellingwerf, K.J, Crielaard, W, Van Aalten, D.M.F. | | Deposit date: | 2003-03-14 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Pas Domains.Common Structure and Common Flexibility
J.Biol.Chem., 278, 2003
|
|
1ODW
 
 | | Native HIV-1 Proteinase | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Thanki, N, Kervinen, J, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
1ODX
 
 | | HIV-1 Proteinase mutant A71T, V82A | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Kervinen, J, Thanki, N, Zdanov, A, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
1ODY
 
 | | HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, HIV-1 PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
1ODZ
 
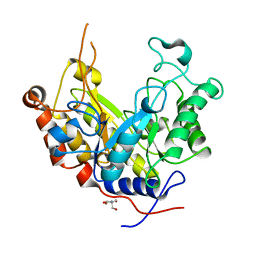 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
1OE0
 
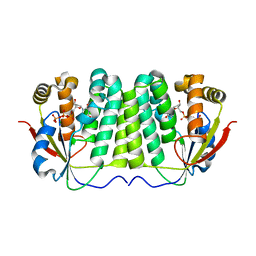 | | CRYSTAL STRUCTURE OF DROSOPHILA DEOXYRIBONUCLEOSIDE KINASE IN COMPLEX WITH DTTP | | Descriptor: | DEOXYRIBONUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-17 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway:Studies of the Drosophila Deoxyribonucleoside Kinase
Biochemistry, 42, 2003
|
|
1OE4
 
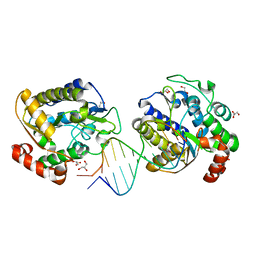 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 5'-D(*CP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*G)-3', 5'-D(*CP*GP*GP*AP*CP*TP*3DR*AP*CP*GP*GP*G)-3', GLYCEROL, ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
1OE5
 
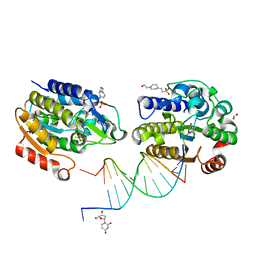 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 2'-DEOXYURIDINE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*CP*3DRP*GP*GP*AP*CP*TP*3DRP*AP*CP*GP*GP*GP)-3', ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
1OE6
 
 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 5'-D(*CP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*G)-3', 5'-D(*CP*GP*GP*AP*CP*TP*3DRP*AP*CP*GP*GP*G)-3', 5-HYDROXYMETHYL URACIL, ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
1OE7
 
 | |
1OE8
 
 | |
1OE9
 
 | | Crystal structure of Myosin V motor with essential light chain-nucleotide-free | | Descriptor: | MYOSIN LIGHT CHAIN 1, SLOW-TWITCH MUSCLE A ISOFORM, MYOSIN VA, ... | | Authors: | Coureux, P.-D, Wells, A.L, Menetrey, J, Yengo, C.M, Morris, C.A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2003-03-21 | | Release date: | 2003-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural State of the Myosin V Motor without Bound Nucleotide
Nature, 425, 2003
|
|
1OEB
 
 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
1OEE
 
 | | YodA from Escherichia coli crystallised with cadmium ions | | Descriptor: | CADMIUM ION, HYPOTHETICAL PROTEIN YODA | | Authors: | David, G, Blondeau, K, Renouard, M, Penel, S, Lewit-Bentley, A. | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Yoda from Escherichia Coli is a Metal-Binding, Lipocalin-Like Protein
J.Biol.Chem., 278, 2003
|
|
