1NK8
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NK9
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER TWO ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP AND DGTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKB
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER THREE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, AND DTTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKC
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER FIVE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, DTTP, AND DATP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKD
 
 | |
1NKE
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A CYTOSINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA POLYMERASE I, DNA PRIMER STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKH
 
 | | Crystal structure of Lactose synthase complex with UDP and Manganese | | Descriptor: | ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2003-01-03 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lactose synthase reveals a large conformational
change in its catalytic component, the beta-1,4-galactosyltransferase
J.Mol.Biol., 310, 2001
|
|
1NKI
 
 | | CRYSTAL STRUCTURE OF THE FOSFOMYCIN RESISTANCE PROTEIN A (FOSA) CONTAINING BOUND PHOSPHONOFORMATE | | Descriptor: | MANGANESE (II) ION, PHOSPHONOFORMIC ACID, POTASSIUM ION, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2003-01-03 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA.
Biochemistry, 43, 2004
|
|
1NKN
 
 | | VISUALIZING AN UNSTABLE COILED COIL: THE CRYSTAL STRUCTURE OF AN N-TERMINAL SEGMENT OF THE SCALLOP MYOSIN ROD | | Descriptor: | S2N51-GCN4 | | Authors: | Li, Y, Brown, J.H, Reshetnikova, L, Blazsek, A, Farkas, L, Nyitray, L, Cohen, C. | | Deposit date: | 2003-01-03 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualization of an unstable coiled coil from the scallop myosin rod
Nature, 424, 2003
|
|
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | Descriptor: | Sialic acid binding Ig-like lectin 7 | | Authors: | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | Deposit date: | 2003-01-03 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NKS
 
 | |
1NKV
 
 | | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13 | | Descriptor: | HYPOTHETICAL PROTEIN yjhP | | Authors: | Kuzin, A, Manor, P, Benach, J, Smith, P, Rost, B, Xiao, R, Montelione, G, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-03 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13
To be published
|
|
1NKZ
 
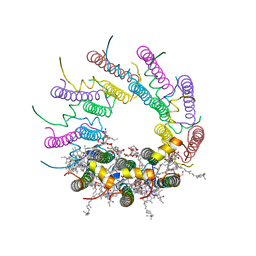 | | Crystal structure of LH2 B800-850 from Rps. acidophila at 2.0 Angstrom resolution | | Descriptor: | BACTERIOCHLOROPHYLL A, BENZAMIDINE, Light-harvesting protein B-800/850, ... | | Authors: | Papiz, M.Z, Prince, S.M, Howard, T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-01-06 | | Release date: | 2003-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure and thermal motion of the B800-850 LH2 complex from Rps. acidophila at 2.0 A resolution and 100K : new structural features and functionally relevant motions.
J.Mol.Biol., 326, 2003
|
|
1NL5
 
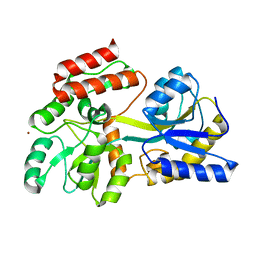 | | Engineered High-affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-01-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
1NL7
 
 | |
1NLC
 
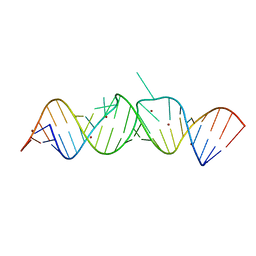 | | HIV-1 DIS(Mal) duplex Zn-soaked | | Descriptor: | HIV-1 DIS(MAL) genomic RNA, ZINC ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2003-01-07 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes.
Nucleic Acids Res., 31, 2003
|
|
1NLD
 
 | | FAB FRAGMENT OF A NEUTRALIZING ANTIBODY DIRECTED AGAINST AN EPITOPE OF GP41 FROM HIV-1 | | Descriptor: | FAB1583 | | Authors: | Davies, C, Beauchamp, J.C, Emery, D, Rawas, A, Muirhead, H. | | Deposit date: | 1996-07-02 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Fab fragment from a neutralizing monoclonal antibody directed against an epitope of gp41 from HIV-1.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1NLF
 
 | | Crystal Structure of DNA Helicase RepA in complex with sulfate at 1.95 A resolution | | Descriptor: | Regulatory protein repA, SULFATE ION | | Authors: | Xu, H, Strater, N, Schroeder, W, Bottcher, C, Ludwig, K, Saenger, W. | | Deposit date: | 2003-01-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of DNA helicase RepA in complex with sulfate at 1.95 A resolution implicates structural changes to an "open" form.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NLI
 
 | | Complex of [E160A-E189A] trichosanthin and adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein alpha-trichosanthin | | Authors: | Shaw, P.C, Wong, K.B, Chan, D.S.B, Williams, R.L. | | Deposit date: | 2003-01-07 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for the interaction of [E160A-E189A]-trichosanthin with adenine.
Toxicon, 41, 2003
|
|
1NLK
 
 | |
1NLN
 
 | | CRYSTAL STRUCTURE OF HUMAN ADENOVIRUS 2 PROTEINASE WITH ITS 11 AMINO ACID COFACTOR AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, Adenain, PVIC | | Authors: | McGrath, W.J, Ding, J, Sweet, R.M, Mangel, W.F. | | Deposit date: | 2003-01-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold
Biochim.Biophys.Acta, 1648, 2003
|
|
1NLQ
 
 | | The crystal structure of Drosophila NLP-core provides insight into pentamer formation and histone binding | | Descriptor: | MAGNESIUM ION, Nucleoplasmin-like protein | | Authors: | Namboodiri, V.M.H, Dutta, S, Akey, I.V, Head, J.F, Akey, C.W. | | Deposit date: | 2003-01-07 | | Release date: | 2003-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of Drosophila NLP-core Provides Insight into Pentamer Formation and Histone Binding
Structure, 11, 2003
|
|
1NLR
 
 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | Descriptor: | ENDO-1,4-BETA-GLUCANASE | | Authors: | Sulzenbacher, G, Dupont, C, Davies, G.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-25 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
1NLS
 
 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | Deposit date: | 1997-01-28 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1NLT
 
 | | The crystal structure of Hsp40 Ydj1 | | Descriptor: | Mitochondrial protein import protein MAS5, Seven residue peptide, ZINC ION | | Authors: | Li, J, Sha, B. | | Deposit date: | 2003-01-07 | | Release date: | 2004-01-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate.
Structure, 11, 2003
|
|
