133D
 
 | | THE CRYSTAL STRUCTURE OF N4-METHYLCYTOSINE.GUANOSIN BASE-PAIRS IN THE SYNTHETIC HEXANUCLEOTIDE D(CGCGM(4)CG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C34)P*G)-3') | | Authors: | Cervi, A.R, Guy, A, Leonard, G.A, Teoule, R, Hunter, W.N. | | Deposit date: | 1993-07-29 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of N4-methylcytosine.guanosine base-pairs in the synthetic hexanucleotide d(CGCGm4CG).
Nucleic Acids Res., 21, 1993
|
|
133L
 
 | |
134L
 
 | |
135L
 
 | |
137D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137L
 
 | |
138D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-HEXAGONAL CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
138L
 
 | |
139L
 
 | |
13GS
 
 | | GLUTATHIONE S-TRANSFERASE COMPLEXED WITH SULFASALAZINE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Ligandin (Non-Substrate) Binding Site of Human Pi Class Glutathione Transferase is Located in the Electrophile Binding Site (H-Site).
J.Mol.Biol., 291, 1999
|
|
140L
 
 | |
141L
 
 | |
142L
 
 | |
143L
 
 | |
144L
 
 | |
145D
 
 | |
145L
 
 | |
146L
 
 | |
147L
 
 | |
148L
 
 | | A COVALENT ENZYME-SUBSTRATE INTERMEDIATE WITH SACCHARIDE DISTORTION IN A MUTANT T4 LYSOZYME | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, BETA-MERCAPTOETHANOL, SUBSTRATE CLEAVED FROM CELL WALL OF ESCHERICHIA COLI, ... | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1993-10-27 | | Release date: | 1994-04-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme.
Science, 262, 1993
|
|
149L
 
 | |
14GS
 
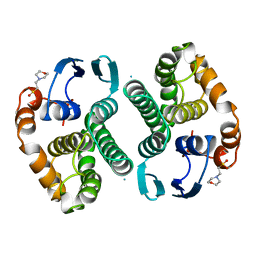 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
150D
 
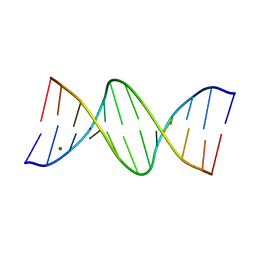 | | GUANINE.1,N6-ETHENOADENINE BASE-PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(EDA)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDA)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Gibson, N.J, Brown, T, Watson, W.P, Hunter, W.N. | | Deposit date: | 1993-12-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine-1,N6-ethenoadenine base pairs in the crystal structure of d(CGCGAATT(epsilon dA)GCG).
Biochemistry, 33, 1994
|
|
150L
 
 | |
151L
 
 | |
