1IBT
 
 | | STRUCTURE OF THE D53,54N MUTANT OF HISTIDINE DECARBOXYLASE AT-170 C | | Descriptor: | HISTIDINE DECARBOXYLASE ALPHA CHAIN, HISTIDINE DECARBOXYLASE BETA CHAIN | | Authors: | Worley, S, Schelp, E, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Proteins, 46, 2002
|
|
1IBU
 
 | | STRUCTURE OF THE D53,54N MUTANT OF HISTIDINE DECARBOXYLASE AT 25 C | | Descriptor: | HISTIDINE DECARBOXYLASE ALPHA CHAIN, HISTIDINE DECARBOXYLASE BETA CHAIN | | Authors: | Worley, S, Schelp, E, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Proteins, 46, 2002
|
|
1IBW
 
 | | STRUCTURE OF THE D53,54N MUTANT OF HISTIDINE DECARBOXYLASE BOUND WITH HISTIDINE METHYL ESTER AT 25 C | | Descriptor: | HISTIDINE DECARBOXYLASE BETA CHAIN, HISTIDINE-METHYL-ESTER, Histidine decarboxylase alpha chain | | Authors: | Worley, S, Schelp, E, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Proteins, 46, 2002
|
|
1IC1
 
 | | THE CRYSTAL STRUCTURE FOR THE N-TERMINAL TWO DOMAINS OF ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Casasnovas, J.M, Stehle, T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1IC2
 
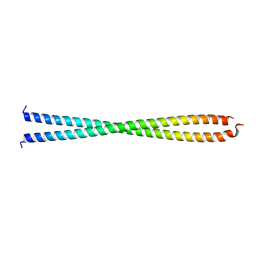 | | DECIPHERING THE DESIGN OF THE TROPOMYOSIN MOLECULE | | Descriptor: | TROPOMYOSIN ALPHA CHAIN, SKELETAL MUSCLE | | Authors: | Brown, J.H, Kim, K.-H, Jun, G, Greenfield, N.J, Dominguez, R, Volkmann, N, Hitchcock-DeGregori, S.E, Cohen, C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the design of the tropomyosin molecule
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IC8
 
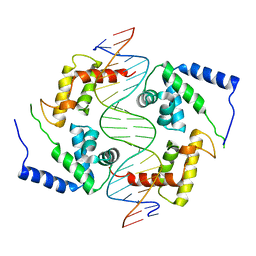 | | HEPATOCYTE NUCLEAR FACTOR 1A BOUND TO DNA : MODY3 GENE PRODUCT | | Descriptor: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*GP*A)-3', 5'-D(*TP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*G)-3', HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Chi, Y.-I, Frantz, J.D, Oh, B.-C, Hansen, L, Dhe-Paganon, S, Shoelson, S.E. | | Deposit date: | 2001-03-30 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Diabetes mutations delineate an
atypical POU domains in HNF1-Alpha
Mol.Cell, 10, 2002
|
|
1ICC
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 OUTER MITOCHONDRIAL MEMBRANE ISOFORM, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X. | | Deposit date: | 2001-03-30 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the differences between rat liver outer mitochondrial membrane cytochrome b5 and microsomal cytochromes b5.
Biochemistry, 40, 2001
|
|
1ICG
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[F]U IN PRESENCE OF IR(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UFP))-3', CHLORIDE ION, IRIDIUM HEXAMMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-03-31 | | Release date: | 2001-04-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ICI
 
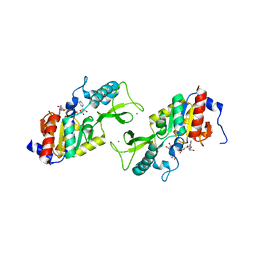 | | CRYSTAL STRUCTURE OF A SIR2 HOMOLOG-NAD COMPLEX | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRANSCRIPTIONAL REGULATORY PROTEIN, SIR2 FAMILY, ... | | Authors: | Min, J, Landry, J, Sternglanz, R, Xu, R.-M. | | Deposit date: | 2001-04-01 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a SIR2 homolog-NAD complex.
Cell(Cambridge,Mass.), 105, 2001
|
|
1ICJ
 
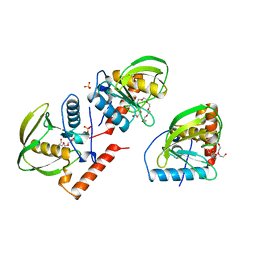 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1ICK
 
 | | LEFT-HANDED Z-DNA HEXAMER DUPLEX D(CGCGCG)2 | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Dauter, Z, Adamiak, D.A. | | Deposit date: | 2001-04-01 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Anomalous signal of phosphorus used for phasing DNA oligomer: importance of data redundancy.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ICM
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, MYRISTIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
1ICN
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICQ
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICS
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICX
 
 | |
1ID0
 
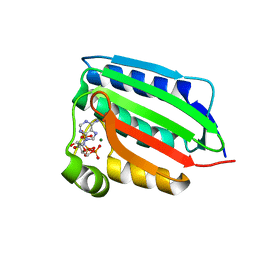 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE BOND CONFORMATION OF PHOQ KINASE DOMAIN | | Descriptor: | MAGNESIUM ION, PHOQ HISTIDINE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Marina, A, Mott, C, Auyzenberg, A, Waldburger, C.D, Hendrickson, W.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism.
J.Biol.Chem., 276, 2001
|
|
1ID1
 
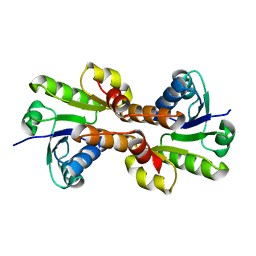 | | CRYSTAL STRUCTURE OF THE RCK DOMAIN FROM E.COLI POTASSIUM CHANNEL | | Descriptor: | PUTATIVE POTASSIUM CHANNEL PROTEIN | | Authors: | Jiang, Y, Pico, A, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel.
Neuron, 29, 2001
|
|
1ID2
 
 | | CRYSTAL STRUCTURE OF AMICYANIN FROM PARACOCCUS VERSUTUS (THIOBACILLUS VERSUTUS) | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Romero, A, Nar, H, Messerschmidt, A. | | Deposit date: | 2001-04-03 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure analysis and refinement at 2.15 A resolution of amicyanin, a type I blue copper protein, from Thiobacillus versutus.
J.Mol.Biol., 236, 1994
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
1ID5
 
 | |
1IDJ
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
1IDK
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
1IDO
 
 | | I-DOMAIN FROM INTEGRIN CR3, MG2+ BOUND | | Descriptor: | INTEGRIN, MAGNESIUM ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-03-12 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18).
Cell(Cambridge,Mass.), 80, 1995
|
|
