4EX5
 
 | |
4EX6
 
 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX7
 
 | | Crystal structure of the alnumycin P phosphatase in complex with free phosphate | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX8
 
 | | Crystal structure of the prealnumycin C-glycosynthase AlnA | | Descriptor: | AlnA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX9
 
 | | Crystal structure of the prealnumycin C-glycosynthase AlnA in complex with ribulose 5-phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AlnA, CALCIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EXA
 
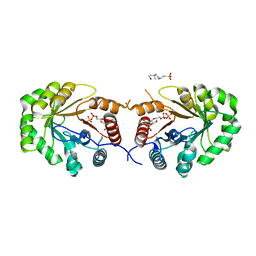 | | Crystal structure of the PA4992, the putative aldo-keto reductase from Pseudomona aeruginosa | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4EXB
 
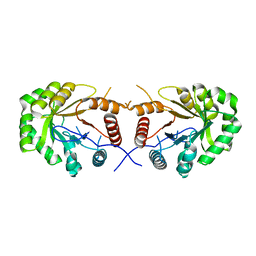 | |
4EXG
 
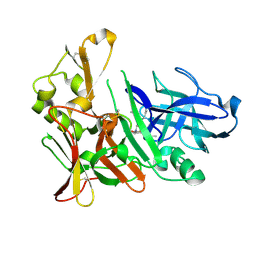 | | Design and synthesis of potent hydroxyethylamine (hea) bace-1 inhibitors | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-4-{[(4S)-6-(2,2-dimethylpropyl)-2,2-dimethyl-3,4-dihydro-2H-thieno[2,3-b]pyran-4-yl]amino}-3-hydroxy-1-phenylbutan-2-yl]acetamide | | Authors: | Borkakoti, N, Lindberg, J, Derbyshire, D. | | Deposit date: | 2012-04-30 | | Release date: | 2012-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of potent hydroxyethylamine (HEA) BACE-1 inhibitors carrying prime side 4,5,6,7-tetrahydrobenzazole and 4,5,6,7-tetrahydropyridinoazole templates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EXH
 
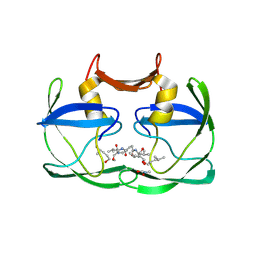 | |
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
4EXK
 
 | | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 from Salmonella enterica | | Descriptor: | Maltose-binding periplasmic protein, uncharacterized protein chimera, TRIETHYLENE GLYCOL, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 form Salmonella enterica
To be Published
|
|
4EXL
 
 | | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphate-binding protein pstS 1 | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A
To be Published
|
|
4EXM
 
 | | The crystal structure of an engineered phage lysin containing the binding domain of pesticin and the killing domain of T4-lysozyme | | Descriptor: | Pesticin, Lysozyme Chimera | | Authors: | Seddiki, N, Noinaj, N, Fairman, J.W, Lukacik, P, Barnard, T.J, Buchanan, S.K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EXN
 
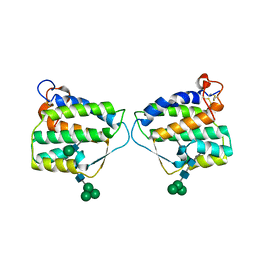 | | Crystal structure of mouse Interleukin-34 | | Descriptor: | Interleukin-34, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
4EXO
 
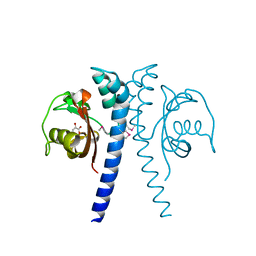 | | Revised, rerefined crystal structure of PDB entry 2QHK, methyl accepting chemotaxis protein | | Descriptor: | Methyl-accepting chemotaxis protein, PYRUVIC ACID | | Authors: | Sweeney, E.G, Henderson, J.N, Goers, J, Wreden, C, Hicks, K.G, Foster, J.K, Parthasarathy, R, Remington, S.J, Guillemin, K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Proposed Mechanism for the pH-Sensing Helicobacter pylori Chemoreceptor TlpB.
Structure, 20, 2012
|
|
4EXP
 
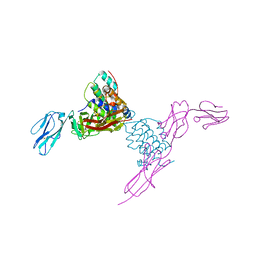 | | Structure of mouse Interleukin-34 in complex with mouse FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-34, Macrophage colony-stimulating factor 1 receptor | | Authors: | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
4EXQ
 
 | |
4EXR
 
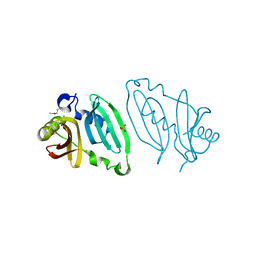 | |
4EXS
 
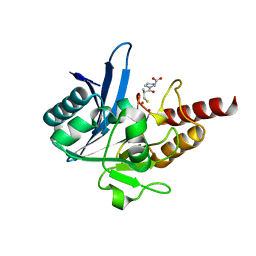 | | Crystal structure of NDM-1 bound to L-captopril | | Descriptor: | Beta-lactamase NDM-1, L-CAPTOPRIL, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EXT
 
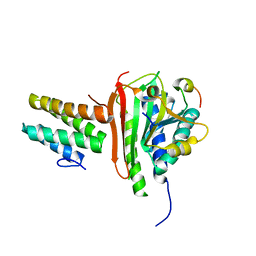 | |
4EXV
 
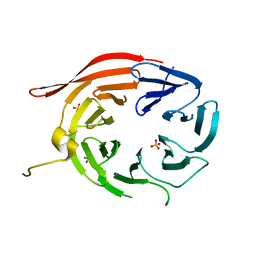 | | Structure of Kluyveromyces lactis Hsv2p | | Descriptor: | SULFATE ION, SVP1-like protein 2 | | Authors: | Baskaran, S, Hurley, J.H. | | Deposit date: | 2012-05-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy.
Mol.Cell, 47, 2012
|
|
4EXW
 
 | |
4EXX
 
 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-05-01 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
4EXY
 
 | |
4EXZ
 
 | |
