1L5T
 
 | | Crystal Structure of a Domain-Opened Mutant (R121D) of the Human Lactoferrin N-lobe Refined From a Merohedrally-Twinned Crystal Form. | | Descriptor: | lactoferrin | | Authors: | Jameson, G.B, Anderson, B.F, Breyer, W.A, Tweedie, J.W, Baker, E.N. | | Deposit date: | 2002-03-07 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a domain-opened mutant (R121D) of the human lactoferrin N-lobe refined from a merohedrally twinned crystal form.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L5U
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 12 base pairs of duplex DNA following addition of a dTTP, a dATP, and a dCTP residue. | | Descriptor: | 5'-D(*GP*A*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*C)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-08 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L5V
 
 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with Glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MALTODEXTRIN PHOSPHORYLASE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L5W
 
 | | Crystal Structure of the Maltodextrin Phosphorylase Complexed with the Products of the Enzymatic Reaction between Glucose-1-phosphate and Maltotetraose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L5X
 
 | | The 2.0-Angstrom resolution crystal structure of a survival protein E (SurE) homolog from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, Survival protein E | | Authors: | Mura, C, Katz, J.E, Clarke, S.G, Eisenberg, D. | | Deposit date: | 2002-03-08 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of an Archaeal Homolog of Survival
Protein E (SurE-alpha): An Acid Phosphatase with Purine
Nucleotide Specificity
J.Mol.Biol., 326, 2003
|
|
1L5Y
 
 | | CRYSTAL STRUCTURE OF MG2+ / BEF3-BOUND RECEIVER DOMAIN OF SINORHIZOBIUM MELILOTI DCTD | | Descriptor: | BERYLLIUM DIFLUORIDE, BERYLLIUM TETRAFLUORIDE ION, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Park, S, Meyer, M, Jones, A.D, Yennawar, H.P, Yennawar, N.H, Nixon, B.T. | | Deposit date: | 2002-03-08 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain
FASEB J., 16, 2002
|
|
1L5Z
 
 | | CRYSTAL STRUCTURE OF THE E121K SUBSTITUTION OF THE RECEIVER DOMAIN OF SINORHIZOBIUM MELILOTI DCTD | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN DCTD, GLYCEROL, SULFATE ION | | Authors: | Park, S, Meyer, M, Jones, A.D, Yennawar, H.P, Yennawar, N.H, Nixon, B.T. | | Deposit date: | 2002-03-08 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain
FASEB J., 16, 2002
|
|
1L60
 
 | |
1L61
 
 | |
1L62
 
 | |
1L63
 
 | |
1L64
 
 | |
1L65
 
 | |
1L66
 
 | |
1L67
 
 | |
1L68
 
 | |
1L69
 
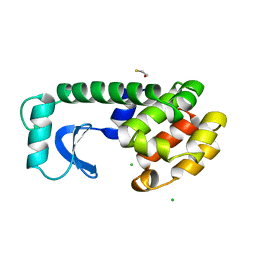 | |
1L6B
 
 | |
1L6I
 
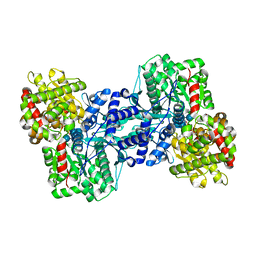 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with the products of the enzymatic reaction between glucose-1-phosphate and maltopentaose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-11 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L6J
 
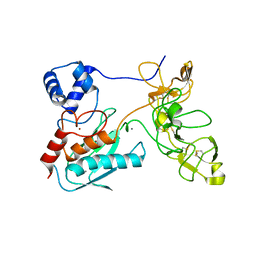 | | Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B). | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, ZINC ION | | Authors: | Elkins, P.A, Ho, Y.S, Smith, W.W, Janson, C.A, D'Alessio, K.J, McQueney, M.S, Cummings, M.D, Romanic, A.M. | | Deposit date: | 2002-03-11 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L6P
 
 | |
1L6R
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1L6W
 
 | | Fructose-6-phosphate aldolase | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL | | Authors: | Thorell, S, Schuermann, M, Sprenger, G.A, Schneider, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family.
J.Mol.Biol., 319, 2002
|
|
1L6X
 
 | | FC FRAGMENT OF RITUXIMAB BOUND TO A MINIMIZED VERSION OF THE B-DOMAIN FROM PROTEIN A CALLED Z34C | | Descriptor: | IMMUNOGLOBULIN GAMMA-1 HEAVY CHAIN CONSTANT REGION, Minimized B-domain of Protein A Z34C, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Idusogie, E.E, Presta, L.G, Santoro-Gazzano, H, Totpal, K, Wong, P.Y, Ultsch, M, Meng, Y.G, Mullkerrin, M.G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc.
J.Immunol., 164, 2000
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
