1KHJ
 
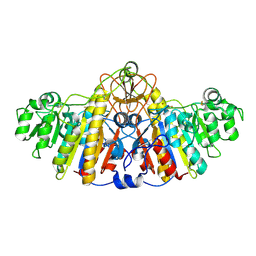 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALUMINUM FLUORIDE, Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHK
 
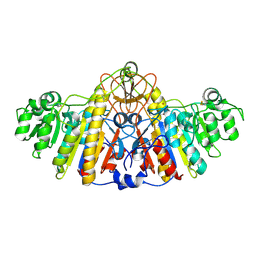 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) | | Descriptor: | Alkaline Phosphatase, MAGNESIUM ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHL
 
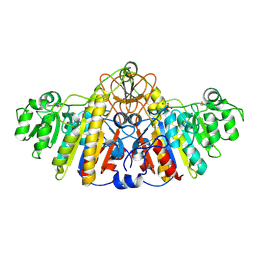 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline Phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHN
 
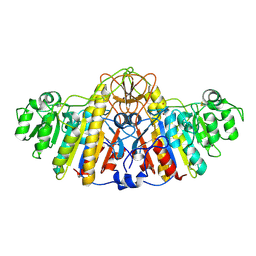 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) ZINC FORM | | Descriptor: | Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHP
 
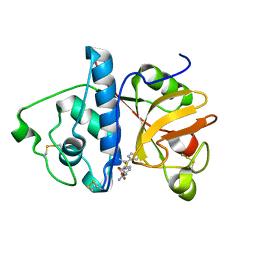 | | Monoclinic form of papain/ZLFG-DAM covalent complex | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
1KHQ
 
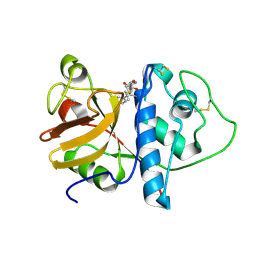 | | ORTHORHOMBIC FORM OF PAPAIN/ZLFG-DAM COVALENT COMPLEX | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
1KHT
 
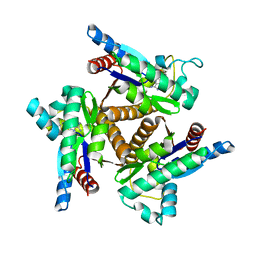 | |
1KHU
 
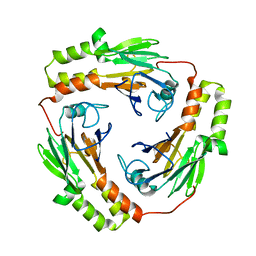 | |
1KHV
 
 | | Crystal Structure of Rabbit Hemorrhagic Disease Virus RNA-dependent RNA polymerase complexed with Lu3+ | | Descriptor: | LUTETIUM (III) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ng, K.K, Cherney, M.M, Vazquez, A.L, Machin, A, Alonso, J.M, Parra, F, James, M.N. | | Deposit date: | 2001-12-01 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase.
J.Biol.Chem., 277, 2002
|
|
1KHW
 
 | | Crystal Structure of Rabbit Hemorrhagic Disease Virus RNA-dependent RNA polymerase complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ng, K.K, Cherney, M.M, Vazquez, A.L, Machin, A, Alonso, J.M, Parra, F, James, M.N. | | Deposit date: | 2001-12-01 | | Release date: | 2002-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase.
J.Biol.Chem., 277, 2002
|
|
1KHY
 
 | |
1KI2
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH GANCICLOVIR | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI3
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH PENCICLOVIR | | Descriptor: | 9-(4-HYDROXY-3-(HYDROXYMETHYL)BUT-1-YL)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Jarvest, R.L, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI4
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOTHIENYLDEOXYURIDINE | | Descriptor: | 5-BROMOTHIENYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI6
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH A 5-IODOURACIL ANHYDROHEXITOL NUCLEOSIDE | | Descriptor: | 1',5'-ANHYDRO-2',3'-DIDEOXY-2'-(5-IODOURACIL-1-YL)-D-ABABINO-HEXITOL, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Herdewijn, P, Ostrowski, T, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI7
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-IODODEOXYURIDINE | | Descriptor: | 5-IODODEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI8
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOVINYLDEOXYURIDINE | | Descriptor: | 5-BROMOVINYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI9
 
 | |
1KIA
 
 | |
1KIB
 
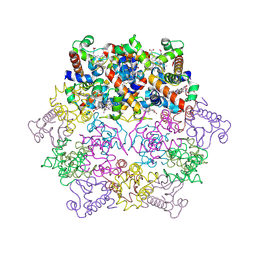 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KIC
 
 | | Inosine-adenosine-guanosine preferring nucleoside hydrolase from Trypanosoma vivax: Asp10Ala mutant in complex with inosine | | Descriptor: | CALCIUM ION, INOSINE, NICKEL (II) ION, ... | | Authors: | Versees, W, Decanniere, K, Van Holsbeke, E, Devroede, N, Steyaert, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzyme-substrate interactions in the purine-specific nucleoside hydrolase from Trypanosoma vivax.
J.Biol.Chem., 277, 2002
|
|
1KIE
 
 | | Inosine-adenosine-guanosine preferring nucleoside hydrolase from Trypanosoma vivax: Asp10Ala mutant in complex with 3-deaza-adenosine | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, NICKEL (II) ION, ... | | Authors: | Versees, W, Decanniere, K, Van Holsbeke, E, Devroede, N, Steyaert, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzyme-substrate interactions in the purine-specific nucleoside hydrolase from Trypanosoma vivax.
J.Biol.Chem., 277, 2002
|
|
1KIJ
 
 | | Crystal structure of the 43K ATPase domain of Thermus thermophilus gyrase B in complex with novobiocin | | Descriptor: | DNA GYRASE SUBUNIT B, FORMIC ACID, NOVOBIOCIN | | Authors: | Lamour, V, Hoermann, L, Jeltsch, J.-M, Oudet, P, Moras, D. | | Deposit date: | 2001-12-03 | | Release date: | 2002-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An open conformation of the Thermus thermophilus gyrase B ATP-binding domain.
J.Biol.Chem., 277, 2002
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
1KIM
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH DEOXYTHYMIDINE | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
