1JBG
 
 | | Crystal Structure of MtaN, the Bacillus subtilis Multidrug Transporter Activator, N-terminus | | Descriptor: | transcription activator of multidrug-efflux transporter genes mta | | Authors: | Godsey, M.H, Neyfakh, A.A, Brennan, R.G. | | Deposit date: | 2001-06-04 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of MtaN, a global multidrug transporter gene activator.
J.Biol.Chem., 276, 2001
|
|
1JBK
 
 | |
1JBM
 
 | |
1JBO
 
 | | The 1.45A Three-Dimensional Structure of c-Phycocyanin from the Thermophylic Cyanobacterium Synechococcus elongatus | | Descriptor: | C-Phycocyanin alpha chain, C-Phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Nield, J, Rizkallah, P.J, Barber, J, Chayen, N.E. | | Deposit date: | 2002-05-02 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The 1.45A three-dimensional structure of C-phycocyanin from the
thermophilic cyanobacterium Synechococcus elongatus
J.STRUCT.BIOL., 141, 2003
|
|
1JBQ
 
 | | STRUCTURE OF HUMAN CYSTATHIONINE BETA-SYNTHASE: A UNIQUE PYRIDOXAL 5'-PHOSPHATE DEPENDENT HEMEPROTEIN | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meier, M, Janosik, M, Kery, V, Kraus, J.P, Burkhard, P. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein.
EMBO J., 20, 2001
|
|
1JBR
 
 | | Crystal Structure of the Ribotoxin Restrictocin and a 31-mer SRD RNA Inhibitor | | Descriptor: | 31-mer SRD RNA analog, 5'-R(*GP*CP*GP*CP*UP*CP*CP*UP*CP*AP*GP*UP*AP*CP*GP*AP*GP*(A23))-3', 5'-R(*GP*GP*AP*AP*CP*CP*GP*GP*AP*GP*CP*GP*C)-3', ... | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBS
 
 | | Crystal structure of ribotoxin restrictocin and a 29-mer SRD RNA analog | | Descriptor: | 29-mer sarcin/ricin domain RNA analog, POTASSIUM ION, restrictocin | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBT
 
 | | CRYSTAL STRUCTURE OF RIBOTOXIN RESTRICTOCIN COMPLEXED WITH A 29-MER SARCIN/RICIN DOMAIN RNA ANALOG | | Descriptor: | 29-MER SARCIN/RICIN DOMAIN RNA ANALOG, POTASSIUM ION, RESTRICTOCIN | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBY
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT LOW PH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1JBZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT HIGH PH | | Descriptor: | 1,2-ETHANEDIOL, GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1JC0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A REDUCED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
1JC1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A OXIDIZED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
1JC4
 
 | | Crystal Structure of Se-Met Methylmalonyl-CoA Epimerase | | Descriptor: | Methylmalonyl-CoA epimerase, SULFATE ION | | Authors: | Mc Carthy, A.A, Baker, H.M, Shewry, S.C, Patchett, M.L, Baker, E.N. | | Deposit date: | 2001-06-07 | | Release date: | 2001-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
Structure, 9, 2001
|
|
1JC5
 
 | | Crystal Structure of Native Methylmalonyl-CoA Epimerase | | Descriptor: | Methylmalonyl-CoA Epimerase, SULFATE ION | | Authors: | Mc Carthy, A.A, Baker, H.M, Shewry, S.C, Patchett, M.L, Baker, E.N. | | Deposit date: | 2001-06-07 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
Structure, 9, 2001
|
|
1JC7
 
 | |
1JCC
 
 | | Crystal Structure of a Novel Alanine-Zipper Trimer at 1.7 A Resolution, V13A,L16A,V20A,L23A,V27A,M30A,V34A mutations | | Descriptor: | MAJOR OUTER MEMBRANE LIPOPROTEIN, ZINC ION | | Authors: | Liu, J, Dai, J, Lu, M. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Zinc-Mediated Helix Capping in A Triple-Helical Protein
Biochemistry, 42, 2003
|
|
1JCD
 
 | | Crystal Structure of a Novel Alanine-Zipper Trimer at 1.3 A Resolution, I6A,L9A,V13A,L16A,V20A,L23A,V27A,M30A,V34A,L48A,M51A mutations | | Descriptor: | MAJOR OUTER MEMBRANE LIPOPROTEIN | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An Alanine-Zipper Structure
Determined by Long Range
Intermolecular Interactions
J.Biol.Chem., 277, 2002
|
|
1JCE
 
 | |
1JCF
 
 | |
1JCG
 
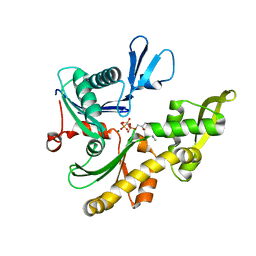 | | MREB FROM THERMOTOGA MARITIMA, AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ROD SHAPE-DETERMINING PROTEIN MREB | | Authors: | van den Ent, F, Amos, L.A, Lowe, J. | | Deposit date: | 2001-06-09 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prokaryotic origin of the actin cytoskeleton.
Nature, 413, 2001
|
|
1JCH
 
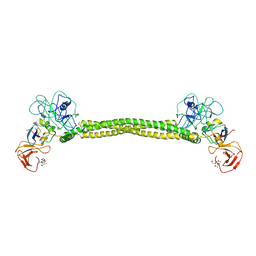 | | Crystal Structure of Colicin E3 in Complex with its Immunity Protein | | Descriptor: | CITRIC ACID, COLICIN E3, COLICIN E3 IMMUNITY PROTEIN, ... | | Authors: | Soelaiman, S, Jakes, K, Wu, N, Li, C, Shoham, M. | | Deposit date: | 2001-06-09 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation.
Mol.Cell, 8, 2001
|
|
1JCJ
 
 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
1JCK
 
 | |
1JCL
 
 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
1JCM
 
 | | TRPC STABILITY MUTANT CONTAINING AN ENGINEERED DISULPHIDE BRIDGE AND IN COMPLEX WITH A CDRP-RELATED SUBSTRATE | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, INDOLE-3-GLYCEROL-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Ivens, A, Mayans, O, Szadkowski, H, Wilmanns, M, Kirschner, K. | | Deposit date: | 2001-06-10 | | Release date: | 2002-06-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stabilization of a (betaalpha)8-barrel protein by an engineered disulfide bridge.
Eur.J.Biochem., 269, 2002
|
|
