6A5J
 
 | | solution NMR Structure of small peptide | | Descriptor: | ILE-LYS-LYS-ILE-LEU-SER-LYS-ILE-LYS-LYS-LEU-LEU-LYS | | Authors: | Feng, L.B, Dong, W.B. | | Deposit date: | 2018-06-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution NMR Structure of antimicrobial peptide L-K6, the analog of temporin-1CEB derived from the skin secretions of Chinese brown frog Rana chensinenesis
To Be Published
|
|
6A7Y
 
 | | Solution structure of an intermolecular leaped V-shape G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*A)-3') | | Authors: | Wan, C.J, Zhang, N. | | Deposit date: | 2018-07-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an asymmetric intermolecular leaped V-shape G-quadruplex: selective recognition of the d(G2NG3NG4) sequence motif by a short linear G-rich DNA probe.
Nucleic Acids Res., 47, 2019
|
|
6A8Y
 
 | | YR26_SDS | | Descriptor: | YR26_SDS | | Authors: | Sinha, S, Bhattacharjya, S. | | Deposit date: | 2018-07-11 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YR26_SDS
To Be Published
|
|
6AAB
 
 | |
6AAS
 
 | | Solution Structure for helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (28-MER) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2025-02-12 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AC7
 
 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
6ACV
 
 | | the solution NMR structure of MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 11 | | Authors: | Li, S.L, Feng, Y.Y, Zhou, Y, Ding, Y.M, Liu, K, Nie, Y, Li, F, Yang, Y.Y. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | the solution NMR structure of MBD domains
To Be Published
|
|
6AFQ
 
 | |
6AGP
 
 | | Structure of Rac1 in the low-affinity state for Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Toyama, Y, Kontani, K, Katada, T, Shimada, I. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational landscape alternations promote oncogenic activities of Ras-related C3 botulinum toxin substrate 1 as revealed by NMR.
Sci Adv, 5, 2019
|
|
6AHZ
 
 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | Descriptor: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | Authors: | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
6AK0
 
 | |
6ALI
 
 | |
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
6ALS
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 4th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALT
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALU
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 8-oxoguanine at the 4th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*CP*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALY
 
 | | Solution structure of yeast Med15 ABD2 residues 277-368 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Tuttle, L.M, Pacheco, D, Warfield, L, Hahn, S, Klevit, R.E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex.
Cell Rep, 22, 2018
|
|
6AMR
 
 | |
6AMV
 
 | |
6AMW
 
 | |
6ANF
 
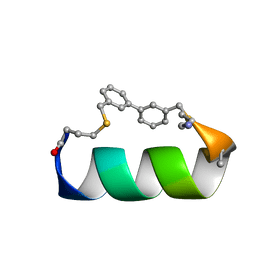 | | Design of a short thermo-stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-08-13 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
6AP5
 
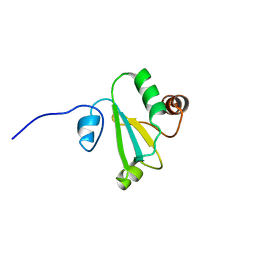 | | H, 13C, and 15N Chemical Shift Assignments and structure of Thioredoxin from Mycobacterium thermoresistibile ATCC 19527 and NCTC 10409 | | Descriptor: | Thioredoxin | | Authors: | Tang, C.T, Yang, F.Y, Varani, G.V, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | H, 13C, and 15N Chemical Shift Assignments and structure of Thioredoxin from Mycobacterium thermoresistibile ATCC 19527 and NCTC 10409
To Be Published
|
|
6ASF
 
 | |
6AST
 
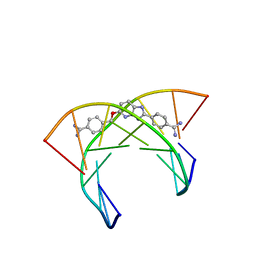 | | NMR and Restrained Molecular Dynamics Determination of the Structure of an Aza-Benzimidazole Derivative Complex with the DNA Minor Groove of an -AAGATA Sequence | | Descriptor: | DNA (5'-D(*CP*CP*AP*AP*GP*AP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*TP*CP*TP*TP*GP*G)-3'), amino(4-{[(2-{4-[amino(iminio)methyl]phenyl}-3H-imidazo[4,5-b]pyridin-5-yl)oxy]methyl}phenyl)methaniminium | | Authors: | Harika, N.K, Germann, M.W, Wilson, W.D. | | Deposit date: | 2017-08-25 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | First Structure of a Designed Minor Groove Binding Heterocyclic Cation that Specifically Recognizes Mixed DNA Base Pair Sequences.
Chemistry, 23, 2017
|
|
