2MFG
 
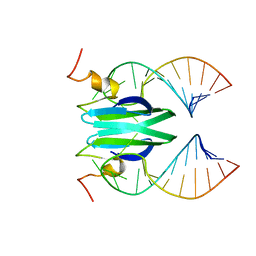 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL4)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL4(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFH
 
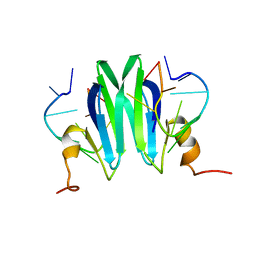 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(36-44)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, RsmZ(36-44) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFI
 
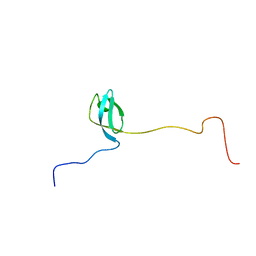 | | Domain 1 of E. coli ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Giraud, P, Crechet, J, Bontems, F, Uzan, M, Sizun, C. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Resonance assignment of the ribosome binding domain of E. coli ribosomal protein S1.
Biomol.Nmr Assign., 9, 2015
|
|
2MFJ
 
 | |
2MFK
 
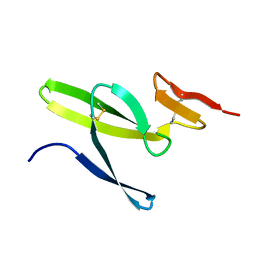 | |
2MFL
 
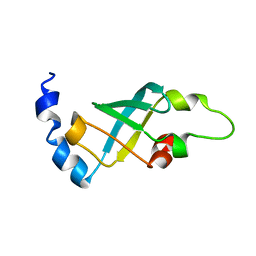 | | Domain 2 of E. coli ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Giraud, P, Crechet, J, Bontems, F, Uzan, M, Sizun, C. | | Deposit date: | 2013-10-12 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Resonance assignment of the ribosome binding domain of E. coli ribosomal protein S1.
Biomol.Nmr Assign., 9, 2015
|
|
2MFM
 
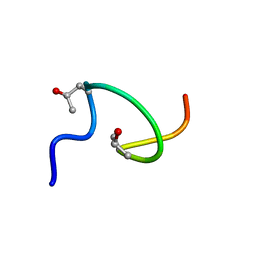 | |
2MFO
 
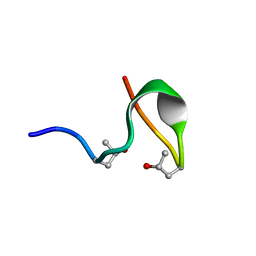 | |
2MFP
 
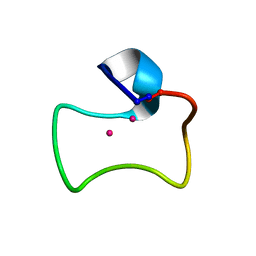 | |
2MFQ
 
 | |
2MFR
 
 | |
2MFS
 
 | |
2MFT
 
 | |
2MFU
 
 | |
2MFV
 
 | |
2MFX
 
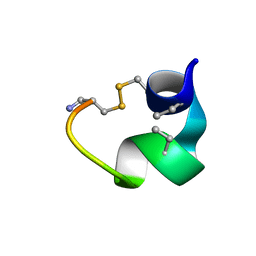 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-cis dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
2MFY
 
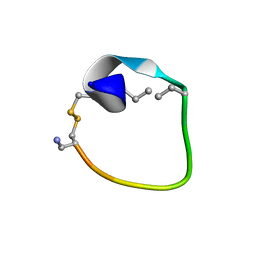 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-trans dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
2MFZ
 
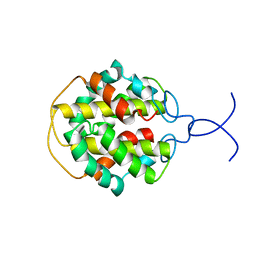 | | NMR structure of C-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Andersson, M, Chen, G, Landreh, M, Nordling, K, Kronqvist, N, Westermark, P, Jornvall, H, Knight, S, Ridderstrale, Y, Holm, L, Meng, Q, Chesler, M, Johansson, J, Rising, A. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains
Plos Biol., 12, 2014
|
|
2MG0
 
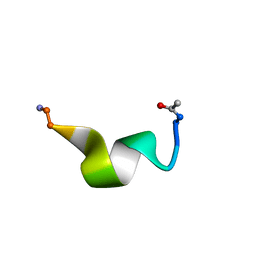 | |
2MG1
 
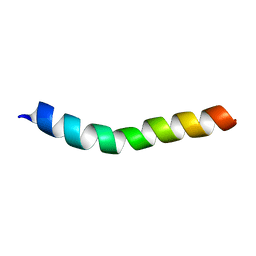 | | NMR assignment and structure of a peptide derived from the trans-membrane region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Apellaniz, B, Huarte, N.L, Nieva, J.L, Jimenez, M.A. | | Deposit date: | 2013-10-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
2MG2
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Apellaniz, B, Huarte, N.L, Nieva, J.L, Jimenez, M.A. | | Deposit date: | 2013-10-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
2MG3
 
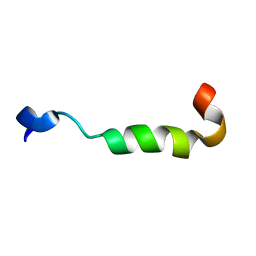 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Serrano, S, Apellaniz, B, Huarte, N, Nieva, J.L, Jimenez, M.A. | | Deposit date: | 2013-10-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
2MG4
 
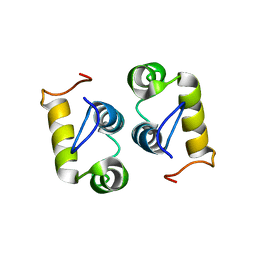 | | Computational design and experimental verification of a symmetric protein homodimer | | Descriptor: | Computational designed homodimer | | Authors: | Mou, Y, Huang, P.S, Hsu, F.C, Huang, S.J, Mayo, S.L. | | Deposit date: | 2013-10-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design and experimental verification of a symmetric protein homodimer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MG5
 
 | |
2MG6
 
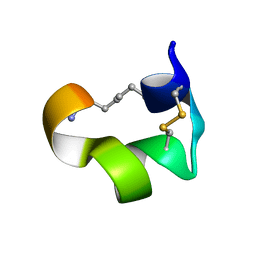 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [3,16]-trans dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-18 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
