5KQB
 
 | |
5KQC
 
 | |
5KQE
 
 | |
5KQJ
 
 | |
5KRW
 
 | | Recognition and targeting mechanisms by chaperones in flagella assembly and operation | | Descriptor: | Flagellar protein FliT,Flagellar hook-associated protein 2 fusion | | Authors: | Khanra, N.K, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition and targeting mechanisms by chaperones in flagellum assembly and operation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KS5
 
 | | Structure of the C-terminal Helical Repeat Domain of Elongation Factor 2 Kinase | | Descriptor: | Eukaryotic elongation factor 2 kinase | | Authors: | Piserchio, A, Will, N, Snyder, I, Ferguson, S.B, Giles, D.H, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-Terminal Helical Repeat Domain of Eukaryotic Elongation Factor 2 Kinase.
Biochemistry, 55, 2016
|
|
5KS6
 
 | |
5KTF
 
 | | Structure of the C-terminal transmembrane domain of scavenger receptor BI (SR-BI) | | Descriptor: | Scavenger receptor class B member 1 | | Authors: | Chadwick, A.C, Peterson, F.C, Volkman, B.F, Sahoo, D. | | Deposit date: | 2016-07-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif.
Structure, 25, 2017
|
|
5KVN
 
 | |
5KVP
 
 | |
5KWO
 
 | |
5KWP
 
 | |
5KWX
 
 | |
5KWZ
 
 | |
5KX0
 
 | |
5KX1
 
 | |
5KX2
 
 | |
5KZO
 
 | |
5L06
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd Position | | Descriptor: | DNA (5'-D(*CP*GP*(5CM)P*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-07-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5L1C
 
 | |
5L2G
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5CM)P*GP*CP*G)-3') | | Authors: | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Johnson, E.C, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-08-01 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5L34
 
 | |
5L3L
 
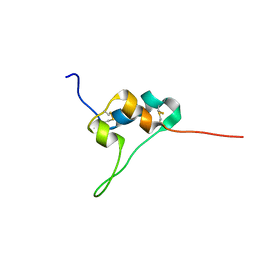 | | D11 bound IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L3M
 
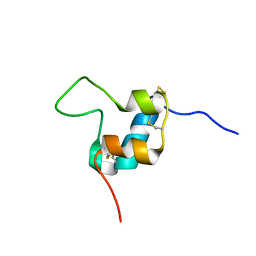 | | D11 bound [S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L3N
 
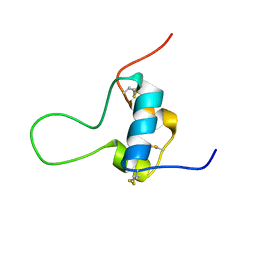 | | D11 bound [N29, S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
