6XRY
 
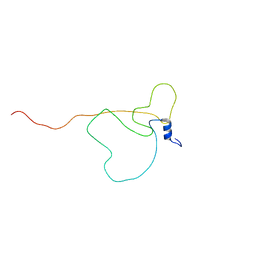 | | Intrinsically disordered bacterial polar organizing protein Z, PopZ, interacts with protein binding partners through an N-terminal Molecular Recognition Feature | | Descriptor: | Polar organizing protein Z | | Authors: | Nordyke, C.T, Ahmed, Y.M, Puterbaugh, R.Z, Bowman, G.R, Varga, K. | | Deposit date: | 2020-07-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Intrinsically Disordered Bacterial Polar Organizing Protein Z, PopZ, Interacts with Protein Binding Partners Through an N-terminal Molecular Recognition Feature.
J.Mol.Biol., 432, 2020
|
|
6XTH
 
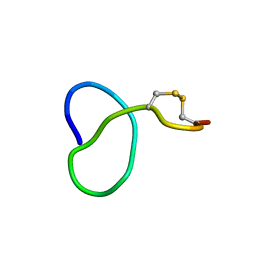 | | NMR solution structure of class IV lasso peptide felipeptin A1 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A1 | | Authors: | Madland, E, Guerrero-Garzon, J.F, Zotchev, S.B, Aachmann, F.L, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6XTI
 
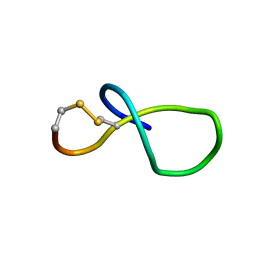 | | NMR solution structure of class IV lasso peptide felipeptin A2 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A2 | | Authors: | Madland, E, Aachmann, F.L, Guerrero-Garzon, J.F, Zotchev, S.B, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6XTT
 
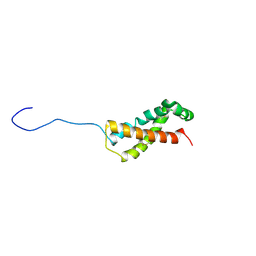 | |
6XV2
 
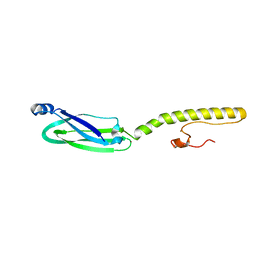 | | Full structure of RYMV P1 protein, derived from crystallographic and NMR data. | | Descriptor: | ZINC ION, p1 | | Authors: | Poignavent, V, Hoh, F, Vignols, F, Demene, H, Yang, Y, Gillet, F.X. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Flexible and Original Architecture of Two Unrelated Zinc Fingers Underlies the Role of the Multitask P1 in RYMV Spread.
J.Mol.Biol., 434, 2022
|
|
6XWI
 
 | |
6XWJ
 
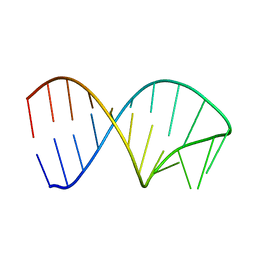 | | Constitutive decay element CDE2 from human 3'UTR | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*CP*UP*AP*AP*UP*AP*UP*UP*UP*AP*GP*GP*CP*AP*CP*C)-3') | | Authors: | Schwalbe, H, Binas, O. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
6XWW
 
 | |
6XXA
 
 | |
6XXB
 
 | |
6XXU
 
 | | Solution NMR structure of the native form of UbcH7 (UBE2L3) | | Descriptor: | Ubiquitin-conjugating enzyme E2 L3 | | Authors: | Marousis, K.D, Seliami, A, Birkou, M, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C,15N backbone and side-chain resonance assignment of the native form of UbcH7 (UBE2L3) through solution NMR spectroscopy.
Biomol.Nmr Assign., 14, 2020
|
|
6XYH
 
 | | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2) | | Descriptor: | AMS3 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2)
To Be Published
|
|
6XYI
 
 | | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3) | | Descriptor: | AMS9.3.1 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3)
TO BE PUBLISHED
|
|
6XYV
 
 | |
6Y06
 
 | |
6Y07
 
 | |
6Y1H
 
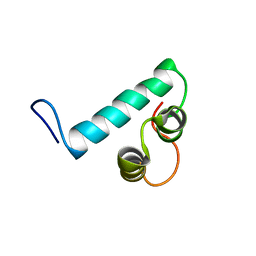 | |
6Y1Q
 
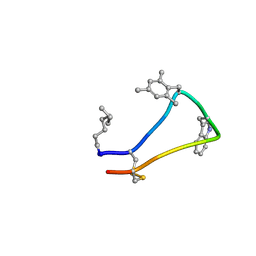 | | Cortistatin analog with improved immunoregulatory activity | | Descriptor: | Analog 5, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Rol, A. | | Deposit date: | 2020-02-13 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of a Cortistatin analogue with immunomodulatory activity in models of inflammatory bowel disease.
Nat Commun, 12, 2021
|
|
6Y3H
 
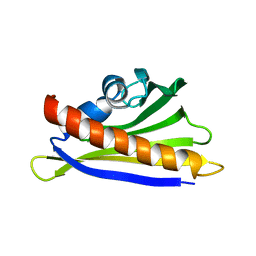 | | NMR solution structure of the hazelnut allergen Cor a 1.0401 | | Descriptor: | Major allergen Cor a 1.0401 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3I
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0402 | | Descriptor: | Major allergen variant Cor a 1.0402 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3K
 
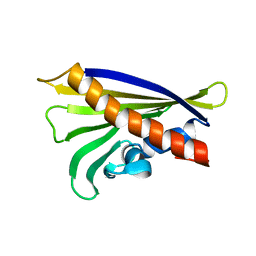 | | NMR solution structure of the hazelnut allergen Cor a 1.0403 | | Descriptor: | Major allergen variant Cor a 1.0403 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3L
 
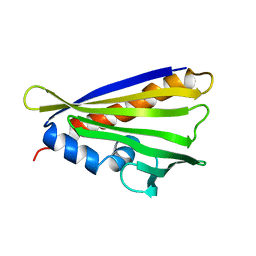 | | NMR solution structure of the hazelnut allergen Cor a 1.0404 | | Descriptor: | Major allergen variant Cor a 1.0404 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y4H
 
 | |
6Y6M
 
 | |
6Y8V
 
 | |
