1GH1
 
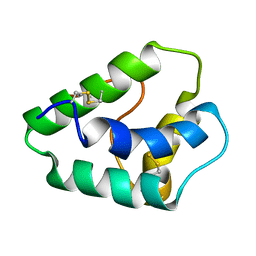 | | NMR STRUCTURES OF WHEAT NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gincel, E, Simorre, J.P, Caille, A, Marion, D, Ptak, M, Vovelle, F. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers.
Eur.J.Biochem., 226, 1994
|
|
1GH5
 
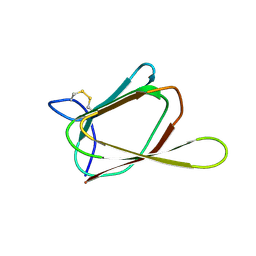 | | ANTIFUNGAL PROTEIN FROM STREPTOMYCES TENDAE TU901, NMR AVERAGE STRUCTURE | | Descriptor: | ANTIFUNGAL PROTEIN | | Authors: | Campos-Olivas, R, Bormann, C, Hoerr, I, Jung, G, Gronenborn, A.M. | | Deposit date: | 2000-11-04 | | Release date: | 2001-03-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TU901.
J.Mol.Biol., 308, 2001
|
|
1GH8
 
 | |
1GH9
 
 | |
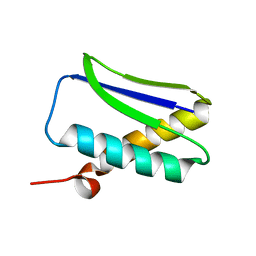
1GHH
 
 | |
1GHJ
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
1GHK
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, 25 STRUCTURES | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
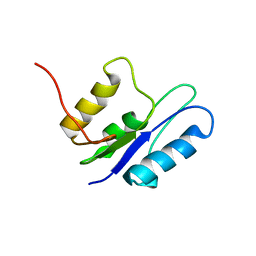
1GHT
 
 | |
1GHU
 
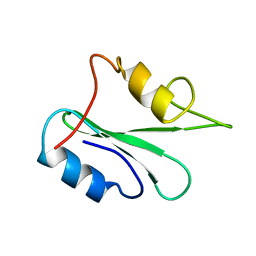 | | NMR solution structure of growth factor receptor-bound protein 2 (GRB2) SH2 domain, 24 structures | | Descriptor: | GRB2 | | Authors: | Thornton, K.H, Mueller, W.T, Mcconnell, P, Zhu, G, Saltiel, A.R, Thanabal, V. | | Deposit date: | 1996-08-05 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the growth factor receptor-bound protein 2 Src homology 2 domain.
Biochemistry, 35, 1996
|
|
1GIB
 
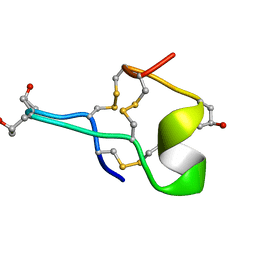 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJX
 
 | | Solution structure of the lipoyl domain of the chimeric dihydrolipoyl dehydrogenase P64K from Neisseria meningitidis | | Descriptor: | PYRUVATE DEHYDROGENASE | | Authors: | Tozawa, K, Broadhurst, R.W, Raine, A.R.C, Fuller, C, Alvarez, A, Guillen, G, Padron, G, Perham, R.N. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Lipoyl Domain of the Chimeric Dihydrolipoyl Dehydrogenase P64K from Neisseria Meningitidis
Eur.J.Biochem., 268, 2001
|
|
1GJZ
 
 | |
1GKS
 
 | | ECTOTHIORHODOSPIRA HALOPHILA CYTOCHROME C551 (REDUCED), NMR, 37 STRUCTURES | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bersch, B, Blackledge, M.J, Meyer, T.E, Marion, D. | | Deposit date: | 1996-07-16 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Ectothiorhodospira halophila ferrocytochrome c551: solution structure and comparison with bacterial cytochromes c.
J.Mol.Biol., 264, 1996
|
|
1GL5
 
 | |
1GM0
 
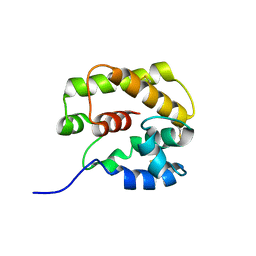 | | A Form of the Pheromone-Binding Protein from Bombyx mori | | Descriptor: | PHEROMONE-BINDING PROTEIN | | Authors: | Horst, R, Damberger, F, Guntert, P, Luginbuhl, P, Nikonova, L, Peng, G, Leal, W.S, Wuthrich, K. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Reveals Novel Intramolecular Regulation Mechanism for Pheromone-Binding and Release
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GNF
 
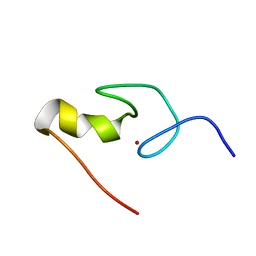 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION FACTOR GATA-1, ZINC ION | | Authors: | Kowalski, K, Czolij, R, King, G.F, Crossley, M, Mackay, J.P. | | Deposit date: | 1998-10-12 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG.
J.Biomol.NMR, 13, 1999
|
|
1GO0
 
 | |
1GO1
 
 | |
1GPS
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-P THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1GPT
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-H THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1GPX
 
 | | C85S GAPDX, NMR, 20 STRUCTURES | | Descriptor: | GALLIUM (III) ION, PUTIDAREDOXIN | | Authors: | Pochapsky, T.C, Kuti, M, Kazanis, S. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a gallium-substituted putidaredoxin mutant: GaPdx C85S.
J.Biomol.NMR, 12, 1998
|
|
1GQ0
 
 | | Solution structure of Antiamoebin I, a membrane channel-forming polypeptide; NMR, 20 structures | | Descriptor: | ANTIAMOEBIN I | | Authors: | Galbraith, T.P, Harris, R, Driscoll, P.C, Wallace, B.A. | | Deposit date: | 2001-11-16 | | Release date: | 2003-01-24 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR studies of antiamoebin, a membrane channel-forming polypeptide.
Biophys. J., 84, 2003
|
|
1GTC
 
 | | HUMAN IMMUNODEFICIENCY VIRUS-1 OKAZAKI FRAGMENT, DNA-RNA CHIMERA, NMR, 11 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*AP*GP*TP*GP*GP*C)-3'), DNA/RNA (5'-R(*GP*CP*CP*A)-D(P*CP*TP*GP*C)-3') | | Authors: | Fedoroff, O.Y, Salazar, M, Reid, B.R. | | Deposit date: | 1996-06-13 | | Release date: | 1996-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural variation among retroviral primer-DNA junctions: solution structure of the HIV-1 (-)-strand Okazaki fragment r(gcca)d(CTGC).d(GCAGTGGC).
Biochemistry, 35, 1996
|
|
