1RZW
 
 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
1S04
 
 | | Solution NMR Structure of Protein PF0455 from Pyrococcus furiosus. Northeast Structural Genomics Consortium Target PfR13 | | Descriptor: | hypothetical protein PF0455 | | Authors: | liu, G, Xiao, R, Sukumaran, D.K, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of The Hypothetical Protein PF0455 From Pyrococcus furiosus: Northeast Structural Genomics Consortium Target PfR13
To be Published
|
|
1S05
 
 | | NMR-validated structural model for oxidized R.palustris cytochrome c556 | | Descriptor: | Cytochrome c-556, HEME C | | Authors: | Bertini, I, Faraone-Mennella, J, Gray, H.B, Luchinat, C, Parigi, G, Winkler, J.R. | | Deposit date: | 2003-12-30 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR-validated structural model for oxidized Rhodopseudomonas palustris cytochrome c(556).
J.Biol.Inorg.Chem., 9, 2004
|
|
1S0T
 
 | | Solution structure of a DNA duplex containing an alpha-anomeric adenosine: insights into substrate recognition by endonuclease IV | | Descriptor: | 5'-D(*Cp*Gp*Tp*Cp*Gp*Tp*Gp*Gp*Ap*C)-3', 5'-D(*Gp*Tp*Cp*Cp*(A3A)p*Cp*Gp*Ap*Cp*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-04 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
1S1N
 
 | | SH3 domain of human nephrocystin | | Descriptor: | Nephrocystin 1 | | Authors: | Le Maire, A, Weber, T, Saunier, S, Antignac, C, Ducruix, A, Dardel, F. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SH3 domain of human nephrocystin and analysis of a mutation-causing juvenile nephronophthisis.
Proteins, 59, 2005
|
|
1S1O
 
 | |
1S24
 
 | | Rubredoxin domain II from Pseudomonas oleovorans | | Descriptor: | CADMIUM ION, Rubredoxin 2 | | Authors: | Perry, A, Tambyrajah, W, Grossmann, J.G, Lian, L.Y, Scrutton, N.S. | | Deposit date: | 2004-01-08 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the two-iron rubredoxin of Pseudomonas oleovorans determined by NMR spectroscopy and solution X-ray scattering and interactions with rubredoxin reductase.
Biochemistry, 43, 2004
|
|
1S2H
 
 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1S34
 
 | | Solution structure of residues 907-929 from Rous Sarcoma Virus | | Descriptor: | 5'-R(*GP*GP*GP*GP*AP*GP*UP*GP*GP*UP*UP*UP*GP*UP*AP*UP*CP*CP*UP*UP*CP*CP*C)-3' | | Authors: | Cabello-Villegas, J, Giles, K.E, Soto, A.M, Yu, P, Beemon, K.L, Wang, Y.X. | | Deposit date: | 2004-01-12 | | Release date: | 2004-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pseudo-5' splice site of a retroviral splicing suppressor.
Rna, 10, 2004
|
|
1S37
 
 | | Accomodation of Mispair-Aligned N3T-Ethyl-N3T DNA Interstrand Crosslink | | Descriptor: | DNA (5'-D(*CP*GP*AP*AP*AP*(TTM)P*TP*TP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*AP*AP*TP*TP*TP*TP*CP*G)-3') | | Authors: | da Silva, M.W, Noronha, A.M, Noll, D.M, Miller, P.S, Colvin, O.M, Gamcsik, M.P. | | Deposit date: | 2004-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Accommodation of mispair aligned N3T-ethyl-N3T DNA interstrand cross link.
Biochemistry, 43, 2004
|
|
1S3A
 
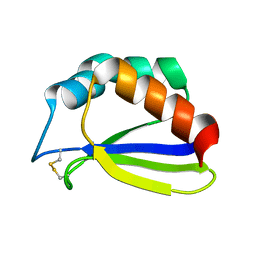 | | NMR Solution Structure of Subunit B8 from Human NADH-Ubiquinone Oxidoreductase Complex I (CI-B8) | | Descriptor: | NADH-ubiquinone oxidoreductase B8 subunit | | Authors: | Brockmann, C, Diehl, A, Rehbein, K, Kuhne, R, Oschkinat, H. | | Deposit date: | 2004-01-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The oxidized subunit B8 from human complex I adopts a thioredoxin fold.
Structure, 12, 2004
|
|
1S4A
 
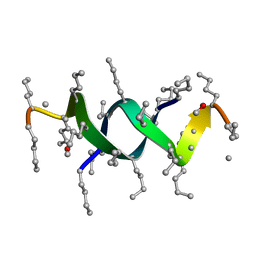 | |
1S4H
 
 | | NMR structure of cross-reactive peptides from L. braziliensis | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
1S4J
 
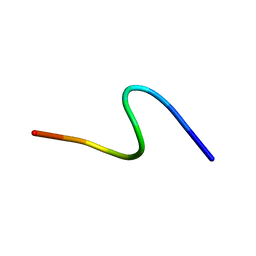 | | NMR structure of cross-reactive peptides from Homo sapiens | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
1S4T
 
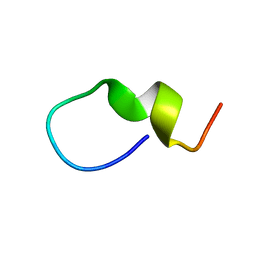 | | Solution structure of synthetic 21mer peptide spanning region 135-155 (in human numbering) of sheep prion protein | | Descriptor: | Major prion protein | | Authors: | Kozin, S.A, Lepage, C, Hui Bon Hoa, G, Rabesona, H, Mazur, A.K, Blond, A, Cheminant, M, Haertle, T, Debey, P, Rebuffat, S. | | Deposit date: | 2004-01-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific recognition between surface loop 2 (132-143) and helix 1 (144-154)
within sheep prion protein from in vitro studies of synthetic peptides
To be Published
|
|
1S5Q
 
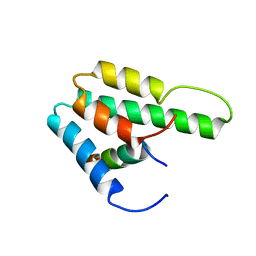 | | Solution Structure of Mad1 SID-mSin3A PAH2 Complex | | Descriptor: | MAD protein, Sin3a protein | | Authors: | Swanson, K.A, Knoepfler, P.S, Huang, K, Kang, R.S, Cowley, S.M, Laherty, C.D, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2004-01-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1S5R
 
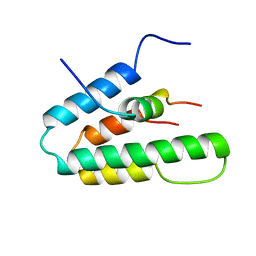 | | Solution Structure of HBP1 SID-mSin3A PAH2 Complex | | Descriptor: | Sin3a protein, high mobility group box transcription factor 1 | | Authors: | Swanson, K.A, Knoepfler, P.S, Huang, K, Kang, R.S, Cowley, S.M, Laherty, C.D, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2004-01-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1S6D
 
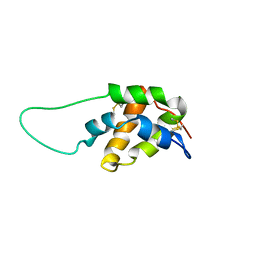 | | Structure in solution of a methionine-rich 2S Albumin protein from Sunflower Seed | | Descriptor: | Albumin 8 | | Authors: | Pantoja-Uceda, D, Bruix, M, Shewry, P.R, Santoro, J, Rico, M. | | Deposit date: | 2004-01-23 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Methionine-Rich 2S Albumin from Sunflower Seeds: Relationship to Its Allergenic and Emulsifying Properties.
Biochemistry, 43, 2004
|
|
1S6I
 
 | |
1S6J
 
 | |
1S6O
 
 | | Solution structure and backbone dynamics of the apo-form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-26 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1S6U
 
 | | Solution structure and backbone dynamics of the Cu(I) form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1S6W
 
 | | Solution Structure of hybrid white striped bass hepcidin | | Descriptor: | Hepcidin | | Authors: | Babon, J.J, Singh, S, Pennington, M.W, Norton, R.S, Westerman, M.E. | | Deposit date: | 2004-01-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections.
J.Biol.Chem., 280, 2005
|
|
1S6X
 
 | | Solution structure of VSTx | | Descriptor: | KvAP CHANNEL | | Authors: | Jung, H.J, Eu, Y.J, Kim, J.I. | | Deposit date: | 2004-01-28 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and lipid membrane partitioning of VSTx1, an inhibitor of the KvAP potassium channel.
Biochemistry, 44, 2005
|
|
1S74
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
