2RN9
 
 | | Solution structure of human apoCox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RNB
 
 | | Solution structure of human Cu(I)Cox17 | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RND
 
 | |
2RNG
 
 | | Solution structure of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Fujitani, N, Osaki, T, Kawabata, S, Nishimura, S, Mizuguchi, M, Aizawa, T, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria
Biochemistry, 47, 2008
|
|
2RNJ
 
 | | NMR Structure of The S. Aureus VraR DNA Binding Domain | | Descriptor: | Response regulator protein vraR | | Authors: | Donaldson, L.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the Staphylococcus aureus Response Regulator VraR DNA Binding Domain Reveals a Dynamic Relationship between It and Its Associated Receiver Domain
Biochemistry, 47, 2008
|
|
2RNK
 
 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2RNM
 
 | | Structure of The HET-s(218-289) prion in its amyloid form obtained by solid-state NMR | | Descriptor: | Small s protein | | Authors: | Wasmer, C, Lange, A, Van Melckebeke, H, Siemer, A, Riek, R, Meier, B.H. | | Deposit date: | 2008-01-24 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core
Science, 319, 2008
|
|
2RNN
 
 | |
2RNO
 
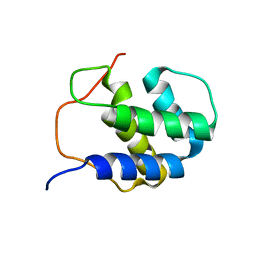 | |
2RNQ
 
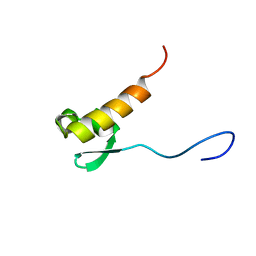 | |
2RNR
 
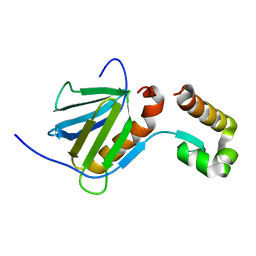 | |
2RNW
 
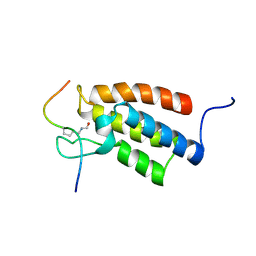 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the Human Transcriptional Co-Activators PCAf and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNX
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNY
 
 | | Complex Structures of CBP Bromodomain with H4 ack20 Peptide | | Descriptor: | CREB-binding protein, Histone H4 | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNZ
 
 | | Solution structure of the presumed chromodomain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
2RO0
 
 | | Solution structure of the knotted tudor domain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
2RO1
 
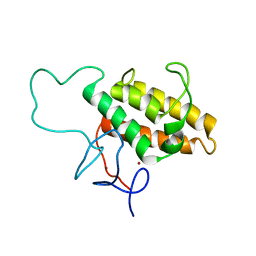 | | NMR Solution Structures of Human KAP1 PHD finger-bromodomain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Zeng, L, Yap, K.L, Ivanov, A.V, Wang, X, Mujtaba, S, Plotnikova, O, Rauscher, F.J. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing
Nat.Struct.Mol.Biol., 15, 2008
|
|
2RO2
 
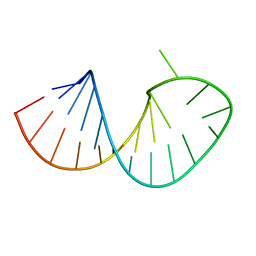 | | Solution structure of domain I of the negative polarity CChMVd hammerhead ribozyme | | Descriptor: | RNA (5'-R(*GP*GP*GP*AP*GP*AP*CP*CP*UP*GP*AP*AP*GP*UP*GP*GP*GP*UP*UP*UP*CP*CP*C)-3') | | Authors: | Gallego, J, Dufour, D, Gago, S, de la Pena, M, Flores, R. | | Deposit date: | 2008-03-05 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads
Nucleic Acids Res., 37, 2009
|
|
2RO3
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator Abh | | Descriptor: | Putative transition state regulator abh | | Authors: | Sullivan, D.M, Bobay, B.G, Douglas, K.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2RO4
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2RO5
 
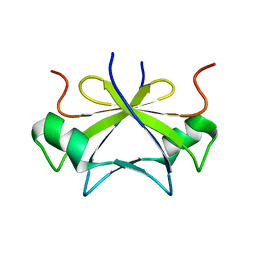 | | RDC-refined solution structure of the N-terminal DNA recognition domain of the Bacillus subtilis transition-state regulator SpoVT | | Descriptor: | Stage V sporulation protein T | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2RO8
 
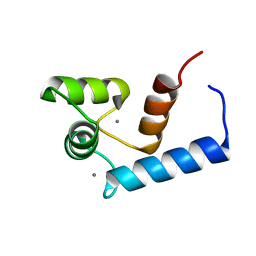 | | Solution structure of calcium bound soybean calmodulin isoform 1 N-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2RO9
 
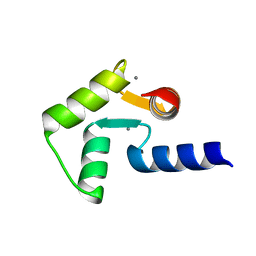 | | Solution structure of calcium bound soybean calmodulin isoform 1 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin-2 | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2ROA
 
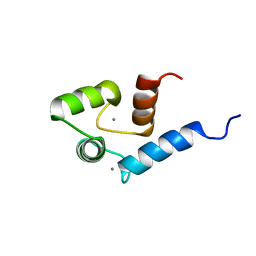 | | Solution structure of calcium bound soybean calmodulin isoform 4 N-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2ROB
 
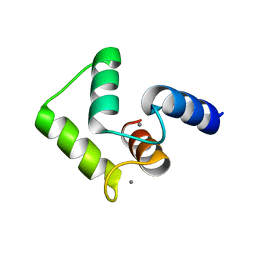 | | Solution structure of calcium bound soybean calmodulin isoform 4 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
