1ELN
 
 | |
1EMN
 
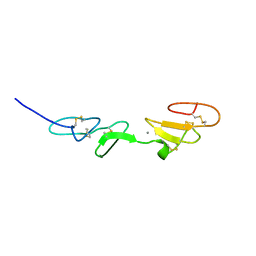 | | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, FIBRILLIN | | Authors: | Downing, A.K, Campbell, I.D, Handford, P.A. | | Deposit date: | 1996-08-05 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Cell(Cambridge,Mass.), 85, 1996
|
|
1EMZ
 
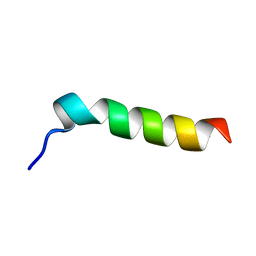 | | SOLUTION STRUCTURE OF FRAGMENT (350-370) OF THE TRANSMEMBRANE DOMAIN OF HEPATITIS C ENVELOPE GLYCOPROTEIN E1 | | Descriptor: | ENVELOPE GLYCOPROTEIN E1 | | Authors: | Op De Beeck, A, Montserret, R, Duvet, S, Cocquerel, L, Cacan, R, Barberot, B, Le Maire, M, Penin, F, Dubuisson, J. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization.
J.Biol.Chem., 275, 2000
|
|
1EO1
 
 | |
1EOT
 
 | |
1EPH
 
 | |
1EPJ
 
 | |
1EQ0
 
 | |
1EQ3
 
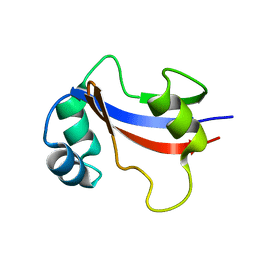 | | NMR STRUCTURE OF HUMAN PARVULIN HPAR14 | | Descriptor: | PEPTIDYL-PROLYL CIS/TRANS ISOMERASE (PPIASE) | | Authors: | Sekerina, E, Rahfeld, U.J, Muller, J, Fischer, G, Bayer, P. | | Deposit date: | 2000-04-02 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of hPar14 reveals similarity to the peptidyl prolyl cis/trans isomerase domain of the mitotic regulator hPin1 but indicates a different functionality of the protein.
J.Mol.Biol., 301, 2000
|
|
1EQX
 
 | | SOLUTION STRUCTURE DETERMINATION AND MUTATIONAL ANALYSIS OF THE PAPILLOMAVIRUS E6-INTERACTING PEPTIDE OF E6AP | | Descriptor: | PAPILLOMAVIRUS E6-ASSOCIATED PROTEIN | | Authors: | Be, X, Hong, Y, Androphy, E.J, Chen, J.J, Baleja, J.D. | | Deposit date: | 2000-04-06 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure determination and mutational analysis of the papillomavirus E6 interacting peptide of E6AP.
Biochemistry, 40, 2001
|
|
1ERC
 
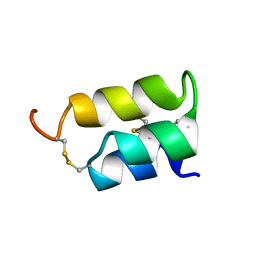 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-1 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-1 | | Authors: | Mronga, S, Luginbuhl, P, Brown, L.R, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-1 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
1ERD
 
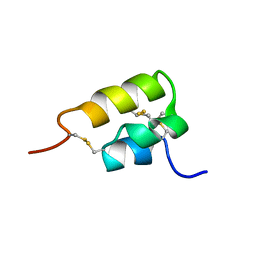 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-2 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-2 | | Authors: | Ottiger, M, Szyperski, T, Luginbuhl, P, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
1ERY
 
 | | PHEROMONE ER-11, NMR | | Descriptor: | PHEROMONE ER-11 | | Authors: | Luginbuhl, P, Wu, J, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-11 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 5, 1996
|
|
1ESH
 
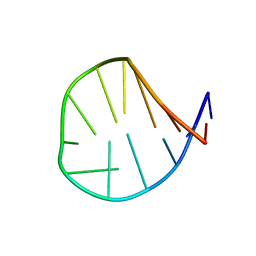 | |
1ESK
 
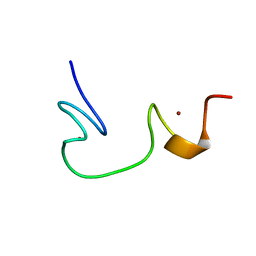 | | SOLUTION STRUCTURE OF NCP7 FROM HIV-1 | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Morellet, N, Demene, H, Teilleux, V, Huynh-Dinh, T, de Rocquigny, H, Fournie-Zaluski, M.-C, Roques, B.P. | | Deposit date: | 2000-04-10 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of (12-53)NCp7 of HIV-1
To be Published
|
|
1ETF
 
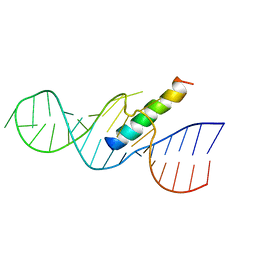 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REV PEPTIDE, REV RESPONSE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1EUB
 
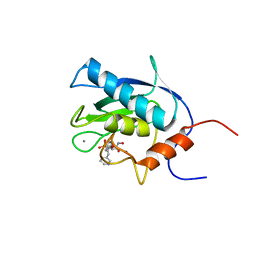 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | Deposit date: | 2000-04-14 | | Release date: | 2001-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
1EV0
 
 | |
1EVB
 
 | |
1EVD
 
 | |
1EW1
 
 | |
1EWW
 
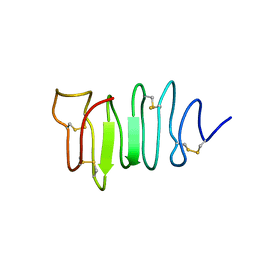 | | SOLUTION STRUCTURE OF SPRUCE BUDWORM ANTIFREEZE PROTEIN AT 30 DEGREES CELSIUS | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Graether, S.P, Kuiper, M.J, Gagne, S.M, Walker, V.K, Jia, Z, Sykes, B.D, Davies, P.L. | | Deposit date: | 2000-04-27 | | Release date: | 2000-07-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect.
Nature, 406, 2000
|
|
1EXE
 
 | |
1EXK
 
 | | SOLUTION STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF THE ESCHERICHIA COLI CHAPERONE PROTEIN DNAJ. | | Descriptor: | DNAJ PROTEIN, ZINC ION | | Authors: | Martinez-Yamout, M, Legge, G.B, Zhang, O, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-05-03 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ.
J.Mol.Biol., 300, 2000
|
|
1EXL
 
 | | STRUCTURE OF AN 11-MER DNA DUPLEX CONTAINING THE CARBOCYCLIC NUCLEOTIDE ANALOG: 2'-DEOXYARISTEROMYCIN | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*(2AR)P*GP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*CP*TP*CP*AP*CP*TP*G)-3') | | Authors: | Smirnov, S, Johnson, F, Marumoto, R, de los Santos, C. | | Deposit date: | 2000-05-03 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an 11-mer DNA duplex containing the carbocyclic nucleotide analog: 2'-deoxyaristeromycin
J.Biomol.Struct.Dyn., 17, 2000
|
|
