1Y8D
 
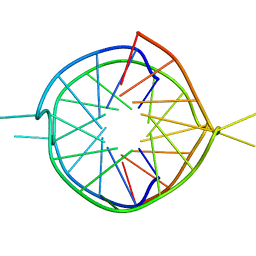 | | Dimeric parallel-stranded tetraplex with 3+1 5' G-tetrad interface, single-residue chain reversal loops and GAG triad in the context of A(GGGG) pentad | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*GP*T)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Ma, J.-B, Faure, A, Andreola, M.-L, Patel, D.J. | | Deposit date: | 2004-12-11 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1Y8F
 
 | |
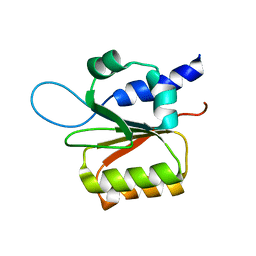
1Y8M
 
 | |
1Y9H
 
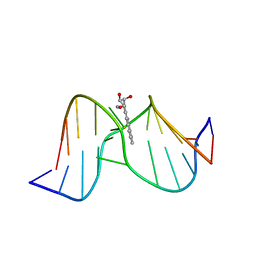 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
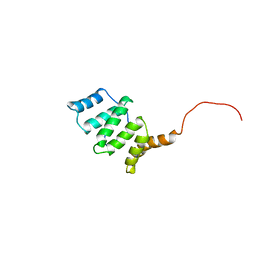
1Y9J
 
 | | Solution structure of the rat Sly1 N-terminal domain | | Descriptor: | Sec1 family domain containing protein 1 | | Authors: | Arac, D, Dulubova, I, Pei, J, Huryeva, I, Grishin, N.V, Rizo, J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Structure of the rSly1 N-terminal Domain Reveals a Conformational Change Induced by Binding to Syntaxin 5.
J.Mol.Biol., 346, 2005
|
|
1Y9O
 
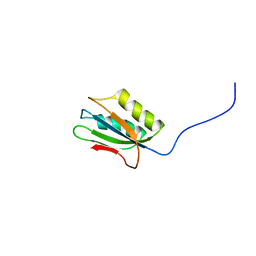 | | 1H NMR Structure of Acylphosphatase from the hyperthermophile Sulfolobus Solfataricus | | Descriptor: | Acylphosphatase | | Authors: | Corazza, A, Rosano, C, Pagano, K, Alverdi, V, Esposito, G, Capanni, C, Bemporad, F, Plakoutsi, G, Stefani, M, Chiti, F, Zuccotti, S, Bolognesi, M, Viglino, P. | | Deposit date: | 2004-12-16 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus
Proteins, 62, 2006
|
|
1Y9X
 
 | |
1YBL
 
 | |
1YBR
 
 | |
1YCM
 
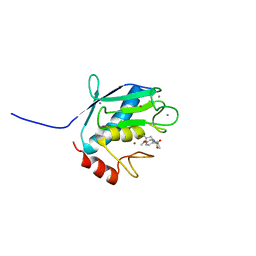 | | Solution Structure of matrix metalloproteinase 12 (MMP12) in the presence of N-Isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.M, Luchinat, C, Mangani, S, Terni, B, Turano, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-12-22 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YCT
 
 | |
1YCW
 
 | |
1YDU
 
 | | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain | | Descriptor: | At5g01610 | | Authors: | Zhao, Q, Cornilescu, C.C, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-26 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain
To be Published
|
|
1YEL
 
 | | Structure of the hypothetical Arabidopsis thaliana protein At1g16640.1 | | Descriptor: | At1g16640 | | Authors: | Peterson, F.C, Waltner, J.K, Lytle, B.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the B3 domain from Arabidopsis thaliana protein At1g16640
Protein Sci., 14, 2005
|
|
1YEZ
 
 | | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30. | | Descriptor: | MM1357 | | Authors: | Rossi, P, Aramini, J.M, Swapna, G.V.T, Huang, Y.P, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30.
To be Published
|
|
1YFB
 
 | |
1YG0
 
 | |
1YG3
 
 | |
1YG4
 
 | |
1YGW
 
 | |
1YH5
 
 | | Solution NMR Structure of Protein yggU from Escherichia coli. Northeast Structural Genomics Consortium Target ER14. | | Descriptor: | ORF, HYPOTHETICAL PROTEIN | | Authors: | Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Wu, M.J, Mills, J.L, Tejero, R.T, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Hypothetical Protein Yggu from E. Coli. Northeast Structural Genomics Consortium Target Er14.
To be Published
|
|
1YHD
 
 | | The solution structure of YGGX from Escherichia Coli | | Descriptor: | UPF0269 protein yggX | | Authors: | Osborne, M.J, Siddiqui, N, Landgraf, D, Pomposiello, P.J, Gehring, K. | | Deposit date: | 2005-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidative stress-related protein YggX from Escherichia coli.
Protein Sci., 14, 2005
|
|
1YHO
 
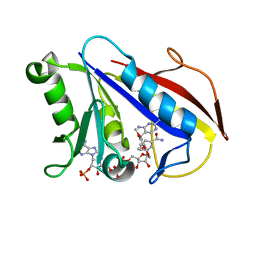 | | Solution structure of human dihydrofolate reductase complexed with trimethoprim and nadph, 25 structures | | Descriptor: | 2,4-DIAMINO-5-(3,4,5-TRIMETHOXY-BENZYL)-PYRIMIDIN-1-IUM, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Polshakov, V.I, Birdsall, B. | | Deposit date: | 2005-01-10 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human dihydrofolate reductase in its complex with trimethoprim and NADPH.
J.Biomol.Nmr, 33, 2005
|
|
1YHP
 
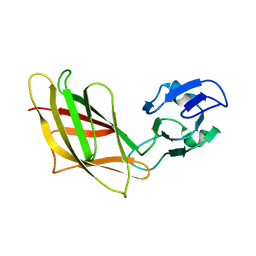 | | Solution Structure of Ca2+-free DdCAD-1 | | Descriptor: | Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
1YJI
 
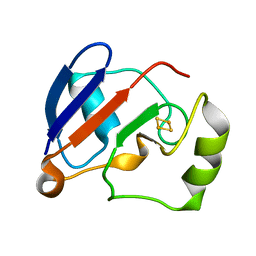 | | RDC-refined Solution NMR structure of reduced putidaredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Jain, N.U, Tjioe, E, Savidor, A, Boulie, J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent structural differences in putidaredoxin derived from homologous structure refinement via residual dipolar couplings.
Biochemistry, 44, 2005
|
|
