1PVE
 
 | |
1PVZ
 
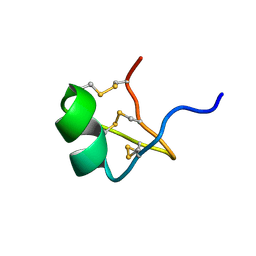 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
1PXE
 
 | |
1PXQ
 
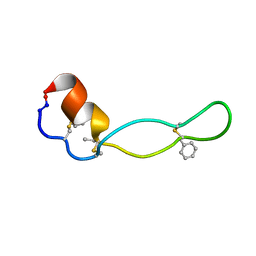 | | Structure of Subtilisin A | | Descriptor: | Subtilisin A | | Authors: | Kawulka, K.E, Sprules, T, McKay, R.T, Mercier, P, Diaper, C.M, Zuber, P, Vederas, J.C. | | Deposit date: | 2003-07-04 | | Release date: | 2004-06-22 | | Last modified: | 2011-10-05 | | Method: | SOLUTION NMR | | Cite: | Structure of subtilisin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to alpha-carbon cross-links: formation and reduction of alpha-thio-alpha-amino acid derivatives
Biochemistry, 43, 2004
|
|
1PYC
 
 | | CYP1 (HAP1) DNA-BINDING DOMAIN (RESIDUES 60-100), NMR, 15 STRUCTURES | | Descriptor: | CYP1, ZINC ION | | Authors: | Timmerman, J, Vuidepot, A.-L, Bontems, F, Lallemand, J.-Y, Gervais, M, Shechter, E, Guiard, B. | | Deposit date: | 1996-02-17 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N resonance assignment and three-dimensional structure of CYP1 (HAP1) DNA-binding domain.
J.Mol.Biol., 259, 1996
|
|
1PYJ
 
 | | Solution Structure of an O6-[4-oxo-4-(3-pyridyl)butyl]guanine adduct in an 11mer DNA duplex | | Descriptor: | 5'-D(*CP*CP*AP*TP*AP*TP*GP*GP*CP*CP*C)-3', 5'-D*GP*GP*GP*CP*CP*AP*TP*AP*TP*GP*G)-3' | | Authors: | Peterson, L.A, Vu, C, Hingerty, B.E, Broyde, S, Cosman, M. | | Deposit date: | 2003-07-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an O6-[4-oxo-4-(3-pyridyl)butyl]guanine adduct in an 11 mer DNA duplex: evidence for formation of a base triplex.
Biochemistry, 42, 2003
|
|
1PYV
 
 | | NMR solution structure of the mitochondrial F1b presequence peptide from Nicotiana plumbaginifolia | | Descriptor: | ATP synthase beta chain, mitochondrial precursor | | Authors: | Moberg, P, Nilsson, S, Stahl, A, Eriksson, A.C, Glaser, E, Maler, L. | | Deposit date: | 2003-07-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the mitochondrial F1beta presequence from Nicotiana plumbaginifolia
J.Mol.Biol., 336, 2004
|
|
1PZQ
 
 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS 2 and DEBS 3: The A domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1PZR
 
 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS2 and DEBS3: the B domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1Q0V
 
 | | Solution Structure of Tandem UIMs of Vps27 | | Descriptor: | hydrophilic protein; has cysteine rich putative zinc finger essential for function; Vps27p | | Authors: | Swanson, K.A, Kang, R.S, Stamenova, S.D, Hicke, L, Radhakrishnan, I. | | Deposit date: | 2003-07-17 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation.
Embo J., 22, 2003
|
|
1Q0W
 
 | | Solution structure of Vps27 amino-terminal UIM-ubiquitin complex | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS27 | | Authors: | Swanson, K.A, Kang, R.S, Stamenova, S.D, Hicke, L, Radhakrishnan, I. | | Deposit date: | 2003-07-17 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vps27 UIM-Ubiquitin Complex Important for Endosomal Sorting and Receptor Downregulation
Embo J., 22, 2003
|
|
1Q10
 
 | |
1Q1V
 
 | |
1Q27
 
 | |
1Q2F
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1Q2I
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1Q2J
 
 | | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA | | Descriptor: | Mu-conotoxin SmIIIA | | Authors: | Keizer, D.W, West, P.J, Lee, E.F, Olivera, B.M, Bulaj, G, Yoshikami, D, Norton, R.S. | | Deposit date: | 2003-07-24 | | Release date: | 2004-02-24 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA.
J.Biol.Chem., 278, 2003
|
|
1Q2K
 
 | | Solution structure of BmBKTx1 a new potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Neurotoxin BmK37 | | Authors: | Cai, Z, Xu, C, Xu, Y, Lu, W, Chi, C.W, Shi, Y, Wu, J. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmBKTx1, a New BK(Ca)(1) Channel Blocker from the Chinese Scorpion Buthus martensi Karsch(,).
Biochemistry, 43, 2004
|
|
1Q2N
 
 | |
1Q2T
 
 | | Solution structure of d(5mCCTCTCC)4 | | Descriptor: | 5'-D(*(MCY)P*CP*TP*CP*TP*CP*C)-3' | | Authors: | Leroy, J.-L. | | Deposit date: | 2003-07-26 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T.T pair intercalation and duplex inter-conversion within i-motif tetramers
J.Mol.Biol., 333, 2003
|
|
1Q2Z
 
 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
1Q38
 
 | | Anastellin | | Descriptor: | Fibronectin | | Authors: | Briknarova, K, Akerman, M.E, Hoyt, D.W, Ruoslahti, E, Ely, K.R. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors
J.Mol.Biol., 332, 2003
|
|
1Q3M
 
 | |
1Q3Y
 
 | | NMR structure of the Cys28His mutant (D form) of the nucleocapsid protein NCp7 of HIV-1. | | Descriptor: | GAG protein, ZINC ION | | Authors: | Ramboarina, S, Druillennec, S, Morellet, N, Bouaziz, S, Roques, B.P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Target specificity of human immunodeficiency virus type 1 NCp7 requires an intact conformation of its CCHC N-terminal zinc finger.
J.Virol., 78, 2004
|
|
1Q3Z
 
 | | NMR structure of the Cys28His mutant (E form) of the nucleocapsid protein NCp7 of HIV-1. | | Descriptor: | GAG protein, ZINC ION | | Authors: | Ramboarina, S, Druillennec, S, Morellet, N, Bouaziz, S, Roques, B.P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Target specificity of human immunodeficiency virus type 1 NCp7 requires an intact conformation of its CCHC N-terminal zinc finger.
J.Virol., 78, 2004
|
|
