2K6R
 
 | | Protein folding on a highly rugged landscape: Experimental observation of glassy dynamics and structural frustration | | Descriptor: | Full Sequence Design 1 Synthetic Superstable | | Authors: | Sadqi, M, de Alba, E, Perez-Jimenez, R, Sanchez-Ruiz, J.M, Munoz, V. | | Deposit date: | 2008-07-18 | | Release date: | 2009-06-16 | | Last modified: | 2016-06-08 | | Method: | SOLUTION NMR | | Cite: | A designed protein as experimental model of primordial folding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K6S
 
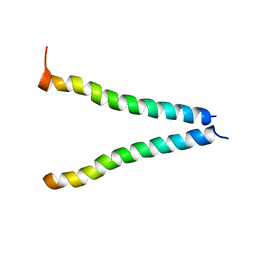 | |
2K6T
 
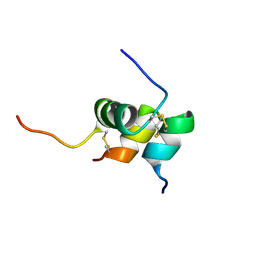 | | Solution structure of the relaxin-like factor | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
2K6U
 
 | | The Solution Structure of a Conformationally Restricted Fully Active Derivative of the Human Relaxin-like Factor (RLF) | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-24 | | Release date: | 2008-12-16 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
2K6V
 
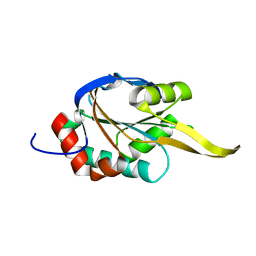 | | Solution structures of apo Sco1 protein from Thermus Thermophilus | | Descriptor: | Putative cytochrome c oxidase assembly protein | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K6W
 
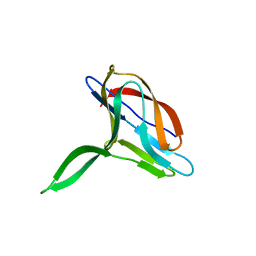 | | Solution structures of apo PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K6X
 
 | | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1- Induced Compacted Structure | | Descriptor: | RNA polymerase sigma factor rpoD | | Authors: | Schwartz, E.C, Shekhtman, A, Dutta, K, Pratt, M.R, Cowburn, D, Darst, S, Muir, T.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1 - Induced Compacted Structure
Chem.Biol., 15, 2008
|
|
2K6Y
 
 | | Solution structures of apo form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K6Z
 
 | | Solution structures of copper loaded form PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K70
 
 | | Solution structures of copper loaded form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K71
 
 | | Structure and dynamics of a DNA GNRA hairpin solved vy high-sensitivity NMR with two independent converging methods, simulated annealing (DYANA) and mesoscopic molecular modelling (BCE/AMBER) | | Descriptor: | 5'-D(*DGP*DCP*DGP*DAP*DAP*DAP*DGP*DC)-3' | | Authors: | Santini, G.P.H, Cognet, J.A.H, Xu, D, Singarapu, K.K, Herve du Penhoat, C.L.M. | | Deposit date: | 2008-07-29 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nucleic acid folding determined by mesoscale modeling and NMR spectroscopy: solution structure of d(GCGAAAGC).
J.Phys.Chem.B, 113, 2009
|
|
2K72
 
 | |
2K73
 
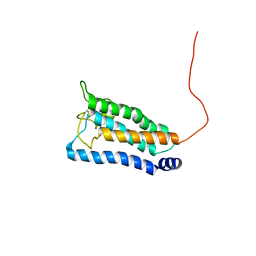 | | Solution NMR structure of integral membrane protein DsbB | | Descriptor: | Disulfide bond formation protein B | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2K74
 
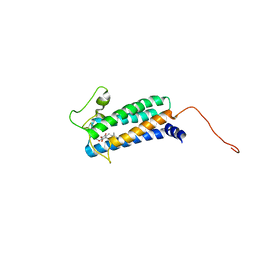 | | Solution NMR structure of DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, UBIQUINONE-2 | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2K75
 
 | | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum. Northeast Structural Genomics Consortium target TaR80b. | | Descriptor: | uncharacterized protein Ta0387 | | Authors: | Ramelot, T.A, Ding, K, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-01 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum.
To be Published
|
|
2K76
 
 | | Solution structure of a paralog-specific Mena binder by NMR | | Descriptor: | pGolemi | | Authors: | Link, N.M, Hunke, C, Eichler, J, Bayer, P. | | Deposit date: | 2008-08-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains.
Biol.Chem., 390, 2009
|
|
2K77
 
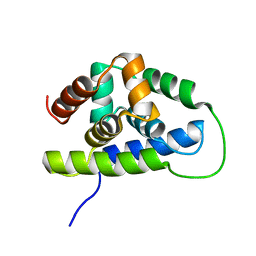 | | NMR solution structure of the Bacillus subtilis ClpC N-domain | | Descriptor: | Negative regulator of genetic competence clpC/mecB | | Authors: | Kojetin, D.J, McLaughlin, P.D, Thompson, R.J, Rance, M, Cavanagh, J. | | Deposit date: | 2008-08-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions.
J.Mol.Biol., 387, 2009
|
|
2K78
 
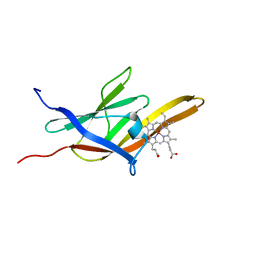 | | Solution Structure of the IsdC NEAT domain bound to Zinc Protoporphyrin | | Descriptor: | Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING ZN | | Authors: | Villareal, V.A, Pilpa, R.M, Robson, S.A, Fadeev, E.A, Clubb, R.T. | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The IsdC Protein from Staphylococcus aureus Uses a Flexible Binding Pocket to Capture Heme.
J.Biol.Chem., 283, 2008
|
|
2K79
 
 | |
2K7A
 
 | |
2K7B
 
 | | NMR structure of Mg2+-bound CaBP1 N-domain | | Descriptor: | Calcium-binding protein 1, MAGNESIUM ION | | Authors: | Ames, J. | | Deposit date: | 2008-08-08 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mg2+-bound CaBP1 N-domain
To be Published
|
|
2K7C
 
 | |
2K7D
 
 | | NMR Structure of Ca2+-bound CaBP1 C-domain | | Descriptor: | CALCIUM ION, Calcium-binding protein 1 | | Authors: | Ames, J. | | Deposit date: | 2008-08-08 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Ca2+-bound CaBP1 C-domain
To be Published
|
|
2K7E
 
 | | NMR structure of the human tRNALys3 bound to the HIV genome Loop I | | Descriptor: | RNA (5'-R(*CP*UP*(SUR)P*UP*UP*AP*AP*(PSU)P*CP*UP*GP*C)-3'), RNA (5'-R(*GP*CP*GP*GP*UP*GP*UP*AP*AP*AP*AP*G)-3') | | Authors: | Bilbille, Y, Vendeix, F.P.A, Guenther, R, Malkiewicz, A, Ariza, X, Vilarrasa, J, Agris, P. | | Deposit date: | 2008-08-09 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs.
Nucleic Acids Res., 37, 2009
|
|
2K7F
 
 | | HADDOCK calculated model of the complex between the BRCT region of RFC p140 and dsDNA | | Descriptor: | 5'-D(P*DCP*DGP*DAP*DCP*DCP*DTP*DCP*DGP*DAP*DGP*DAP*DTP*DCP*DA)-3', 5'-D(P*DCP*DTP*DCP*DGP*DAP*DGP*DGP*DTP*DCP*DG)-3', Replication factor C subunit 1 | | Authors: | Kobayashi, M, Ab, E, Bonvin, A, Siegal, G. | | Deposit date: | 2008-08-10 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex.
J.Biol.Chem., 285, 2010
|
|
