6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | 分子名称: | De novo beta protein | | 著者 | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | 登録日 | 2018-07-19 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6E5H
 
 | |
6E5I
 
 | |
6E5J
 
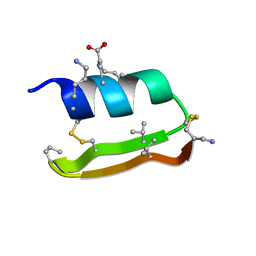 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, beta3 helix, N-methyl hairpin | | 分子名称: | Designed peptide NC_HEE_D1: Aib turn, beta3 helix, N-methyl hairpin mutant | | 著者 | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | 登録日 | 2018-07-20 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
6E5K
 
 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, Aib helix, N-methyl hairpin | | 分子名称: | Designed peptide NC_HEE_D1: Aib turn, Aib helix, N-methyl hairpin mutant | | 著者 | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | 登録日 | 2018-07-20 | | 公開日 | 2018-11-21 | | 最終更新日 | 2020-01-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
6E5N
 
 | |
6E83
 
 | |
6E86
 
 | |
6E8W
 
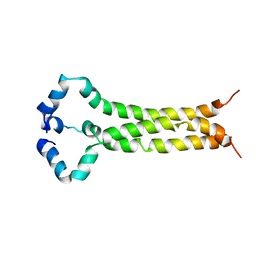 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | 分子名称: | Envelope glycoprotein gp160 | | 著者 | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | 登録日 | 2018-07-31 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E98
 
 | |
6E9M
 
 | |
6ED9
 
 | |
6EE9
 
 | |
6EFE
 
 | |
6EHZ
 
 | |
6EKA
 
 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | 分子名称: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | 著者 | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | 登録日 | 2017-09-25 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6EMO
 
 | |
6EMP
 
 | |
6EMQ
 
 | |
6EMR
 
 | |
6ENA
 
 | |
6EQY
 
 | |
6ER0
 
 | |
6ERL
 
 | |
6ES5
 
 | |
