108D
 
 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | 分子名称: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | 著者 | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | 登録日 | 1995-01-31 | | 公開日 | 1995-06-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
149D
 
 | |
170D
 
 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | 分子名称: | DNA/RNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*T)-R(P*CAR)-D(P*GP*CP*G)-3') | | 著者 | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | 登録日 | 1994-03-14 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
171D
 
 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | 登録日 | 1994-03-14 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
17RA
 
 | |
1A03
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF CA2+-BOUND CALCYCLIN: IMPLICATIONS FOR CA2+-SIGNAL TRANSDUCTION BY S100 PROTEINS, NMR, 20 STRUCTURES | | 分子名称: | CALCYCLIN (RABBIT, CA2+) | | 著者 | Sastry, M, Ketchem, R.R, Crescenzi, O, Weber, C, Lubienski, M.J, Hidaka, H, Chazin, W.J. | | 登録日 | 1997-12-08 | | 公開日 | 1999-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of Ca(2+)-bound calcyclin: implications for Ca(2+)-signal transduction by S100 proteins.
Structure, 6, 1998
|
|
1A1P
 
 | | COMPSTATIN, NMR, 21 STRUCTURES | | 分子名称: | COMPSTATIN | | 著者 | Morikis, D, Assa-Munt, N, Sahu, A, Lambris, J.D. | | 登録日 | 1997-12-12 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Compstatin, a potent complement inhibitor.
Protein Sci., 7, 1998
|
|
1A1T
 
 | | STRUCTURE OF THE HIV-1 NUCLEOCAPSID PROTEIN BOUND TO THE SL3 PSI-RNA RECOGNITION ELEMENT, NMR, 25 STRUCTURES | | 分子名称: | NUCLEOCAPSID PROTEIN, SL3 STEM-LOOP RNA, ZINC ION | | 著者 | De Guzman, R.N, Wu, Z.R, Stalling, C.C, Pappalardo, L, Borer, P.N, Summers, M.F. | | 登録日 | 1997-12-15 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element.
Science, 279, 1998
|
|
1A24
 
 | | SOLUTION NMR STRUCTURE OF REDUCED DSBA FROM ESCHERICHIA COLI, FAMILY OF 20 STRUCTURES | | 分子名称: | DSBA | | 著者 | Schirra, H.J, Renner, C, Czisch, M, Huber-Wunderlich, M, Holak, T.A, Glockshuber, R. | | 登録日 | 1998-01-15 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of reduced DsbA from Escherichia coli in solution.
Biochemistry, 37, 1998
|
|
1A2I
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | 分子名称: | CYTOCHROME C3, HEME C | | 著者 | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | 登録日 | 1998-01-05 | | 公開日 | 1998-07-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
1A2S
 
 | | THE SOLUTION NMR STRUCTURE OF OXIDIZED CYTOCHROME C6 FROM THE GREEN ALGA MONORAPHIDIUM BRAUNII, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | CYTOCHROME C6, HEME C | | 著者 | Banci, L, Bertini, I, De La Rosa, M.A, Koulougliotis, D, Navarro, J.A, Walter, O. | | 登録日 | 1998-01-10 | | 公開日 | 1998-04-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of oxidized cytochrome c6 from the green alga Monoraphidium braunii.
Biochemistry, 37, 1998
|
|
1A3P
 
 | | ROLE OF THE 6-20 DISULFIDE BRIDGE IN THE STRUCTURE AND ACTIVITY OF EPIDERMAL GROWTH FACTOR, NMR, 20 STRUCTURES | | 分子名称: | EPIDERMAL GROWTH FACTOR | | 著者 | Barnham, K, Torres, A, Alewood, D, Alewood, P, Domagala, T, Nice, E, Norton, R. | | 登録日 | 1998-01-22 | | 公開日 | 1998-07-29 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Role of the 6-20 disulfide bridge in the structure and activity of epidermal growth factor.
Protein Sci., 7, 1998
|
|
1A4D
 
 | | LOOP D/LOOP E ARM OF ESCHERICHIA COLI 5S RRNA, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | RNA (5'-R(*GP*GP*CP*CP*GP*AP*UP*GP*GP*UP*AP*GP*UP*GP*UP*GP*GP*GP*GP*UP*C)-3'), RNA (5'-R(P*UP*CP*CP*CP*CP*AP*UP*GP*CP*GP*AP*GP*AP*GP*UP*AP*GP*GP*CP*C)-3') | | 著者 | Dallas, A, Moore, P.B. | | 登録日 | 1998-01-29 | | 公開日 | 1998-04-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The loop E-loop D region of Escherichia coli 5S rRNA: the solution structure reveals an unusual loop that may be important for binding ribosomal proteins.
Structure, 5, 1997
|
|
1A51
 
 | |
1A57
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF A HELIX-LESS VARIANT OF INTESTINAL FATTY ACID BINDING PROTEIN, NMR, 20 STRUCTURES | | 分子名称: | INTESTINAL FATTY ACID-BINDING PROTEIN | | 著者 | Steele, R.A, Emmert, D.A, Kao, J, Hodsdon, M.E, Frieden, C, Cistola, D.P. | | 登録日 | 1998-02-20 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of a helix-less variant of intestinal fatty acid-binding protein.
Protein Sci., 7, 1998
|
|
1A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | 分子名称: | TUMOR SUPPRESSOR P16INK4A | | 著者 | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | 登録日 | 1998-02-13 | | 公開日 | 1999-08-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1A5J
 
 | | CHICKEN B-MYB DNA BINDING DOMAIN, REPEAT 2 AND REPEAT3, NMR, 32 STRUCTURES | | 分子名称: | B-MYB | | 著者 | Mcintosh, P.B, Carr, M.D, Wollborn, U, Frenkiel, T.A, Feeney, J, Mccormick, J.E, Klempnauer, K.H. | | 登録日 | 1998-02-16 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the B-Myb DNA-binding domain: a possible role for conformational instability of the protein in DNA binding and control of gene expression.
Biochemistry, 37, 1998
|
|
1A60
 
 | | NMR STRUCTURE OF A CLASSICAL PSEUDOKNOT: INTERPLAY OF SINGLE-AND DOUBLE-STRANDED RNA, 24 STRUCTURES | | 分子名称: | TYMV PSEUDOKNOT | | 著者 | Kolk, M.H, Van Der Graaf, M, Wijmenga, S.S, Pleij, C.W.A, Heus, H.A, Hilbers, C.W. | | 登録日 | 1998-03-04 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA.
Science, 280, 1998
|
|
1A6X
 
 | | STRUCTURE OF THE APO-BIOTIN CARBOXYL CARRIER PROTEIN (APO-BCCP87) OF ESCHERICHIA COLI ACETYL-COA CARBOXYLASE, NMR, 49 STRUCTURES | | 分子名称: | APO-BIOTIN CARBOXYL CARRIER PROTEIN OF ACETYL-COA CARBOXYLASE | | 著者 | Yao, X, Wei, D, Soden Junior, C, Summers, M.F, Beckett, D. | | 登録日 | 1998-03-04 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the carboxy-terminal fragment of the apo-biotin carboxyl carrier subunit of Escherichia coli acetyl-CoA carboxylase.
Biochemistry, 36, 1997
|
|
1A7F
 
 | |
1A7I
 
 | | AMINO-TERMINAL LIM DOMAIN FROM QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | QCRP2 (LIM1), ZINC ION | | 著者 | Kontaxis, G, Konrat, R, Kraeutler, B, Weiskirchen, R, Bister, K. | | 登録日 | 1998-03-15 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and intramodular dynamics of the amino-terminal LIM domain from quail cysteine- and glycine-rich protein CRP2.
Biochemistry, 37, 1998
|
|
1A7M
 
 | | LEUKAEMIA INHIBITORY FACTOR CHIMERA (MH35-LIF), NMR, 20 STRUCTURES | | 分子名称: | LEUKEMIA INHIBITORY FACTOR | | 著者 | Hinds, M.G, Maurer, T, Zhang, J.-G, Nicola, N.A, Norton, R.S. | | 登録日 | 1998-03-16 | | 公開日 | 1999-04-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of leukemia inhibitory factor.
J.Biol.Chem., 273, 1998
|
|
1A8N
 
 | | SOLUTION STRUCTURE OF A NA+ CATION STABILIZED DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS FORMED BY G-G-G-C REPEATS OBSERVED IN AAV AND HUMAN CHROMOSOME 19, NMR, 8 STRUCTURES | | 分子名称: | DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS | | 著者 | Kettani, A, Bouaziz, S, Gorin, A, Zhao, H, Jones, R, Patel, D.J. | | 登録日 | 1998-03-27 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA.
J.Mol.Biol., 282, 1998
|
|
1A8W
 
 | |
1A93
 
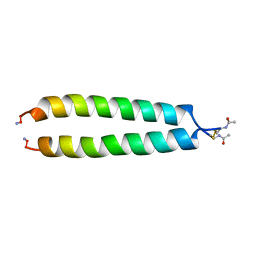 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | 著者 | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | 登録日 | 1998-04-15 | | 公開日 | 1998-10-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
