6ZSO
 
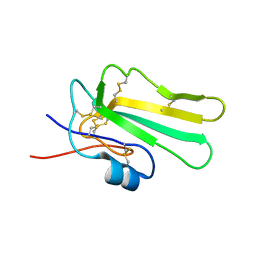 | | Solution structure of the water-soluble LU-domain of human Lypd6b protein | | Descriptor: | Ly6/PLAUR domain-containing protein 6B | | Authors: | Tsarev, A.V, Kulbatskii, D.S, Paramonov, A.S, Lyukmanova, E.N, Shenkarev, Z.O. | | Deposit date: | 2020-07-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors.
Int J Mol Sci, 21, 2020
|
|
6ZSS
 
 | |
6ZTE
 
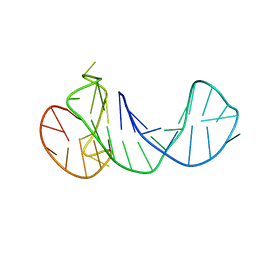 | |
6ZTG
 
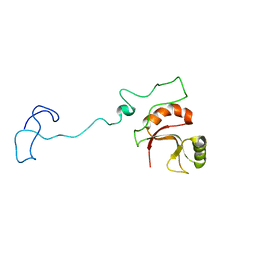 | | Spor protein DedD | | Descriptor: | Cell division protein DedD | | Authors: | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
6ZUY
 
 | | Human TFIIS N-terminal domain (TND) | | Descriptor: | Transcription elongation factor A protein 1 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZUZ
 
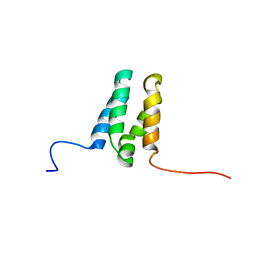 | |
6ZV0
 
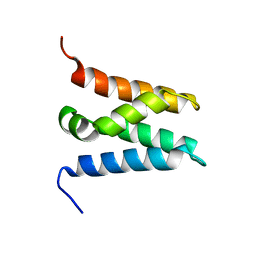 | | TFIIS N-terminal domain (TND) from human LEDGF/p75 | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV1
 
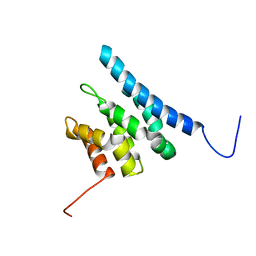 | | TFIIS N-terminal domain (TND) from human IWS1 | | Descriptor: | Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV2
 
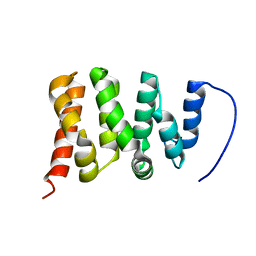 | | TFIIS N-terminal domain (TND) from human PPP1R10 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV3
 
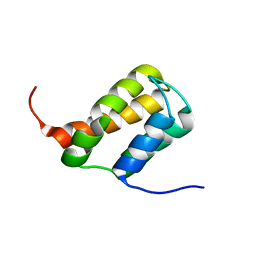 | | TFIIS N-terminal domain (TND) from human MED26 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 26 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV4
 
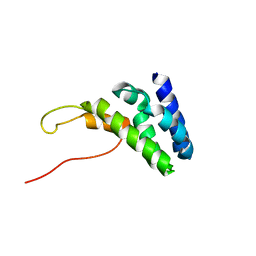 | | Human TFIIS N-terminal domain in complex with IWS1 | | Descriptor: | Transcription elongation factor A protein 1,Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZX6
 
 | |
6ZX7
 
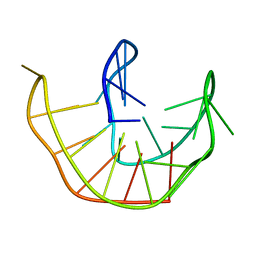 | |
6ZXP
 
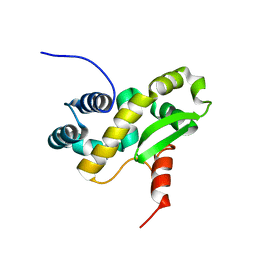 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20 fused to a short peptide from the viral DNA polymerase E9. | | Descriptor: | DNA polymerase processivity factor component A20,DNA polymerase processivity factor component E9 | | Authors: | Bersch, B, Tarbouriech, N, Burmeister, W, Iseni, F. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
6ZYC
 
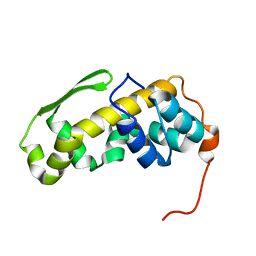 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20. | | Descriptor: | DNA polymerase processivity factor component A20 | | Authors: | Bersch, B, Iseni, F, Burmeister, W, Tarbouriech, N. | | Deposit date: | 2020-07-31 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
6ZYG
 
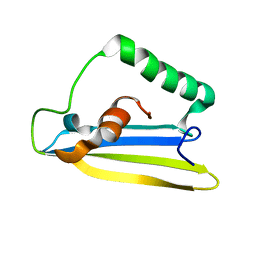 | | Emfourin (M4in) from Serratia proteamaculans, M4 family peptidase inhibitor | | Descriptor: | Protealysin-associated protein | | Authors: | Bozin, T.N, Chukhontseva, K.N, Konarev, P.V, Bocharov, E.V, Demidyuk, I.V. | | Deposit date: | 2020-07-31 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | NMR structure of emfourin, a novel metalloprotease inhibitor
To Be Published
|
|
6ZZE
 
 | |
6ZZF
 
 | |
7A05
 
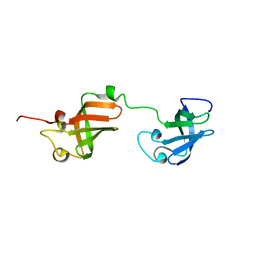 | | NMR structure of D3-D4 domains of Vibrio vulnificus ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Qureshi, N.S, Matzel, T, Cetiner, E.C, Schnieders, S, Jonker, H.R.A, Schwalbe, H, Fuertig, B. | | Deposit date: | 2020-08-06 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Vibrio vulnificus ribosomal protein S1 domains D3 and D4 provides insights into molecular recognition of single-stranded RNAs.
Nucleic Acids Res., 49, 2021
|
|
7A0I
 
 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 7.2 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7A0O
 
 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 5.5 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
7A3Y
 
 | |
7A4L
 
 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | Descriptor: | IRON/SULFUR CLUSTER, PioC | | Authors: | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
7A58
 
 | |
