2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2MVY
 
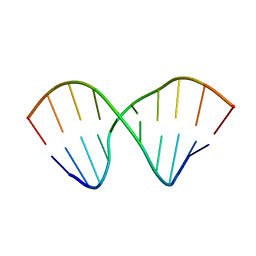 | |
2MVZ
 
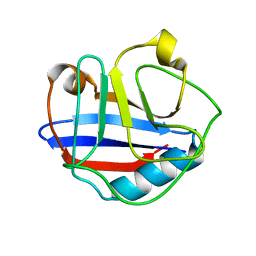 | | Solution Structure for Cyclophilin A from Geobacillus Kaustophilus | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Holliday, M.J, Isern, N.G, Geoffrey, A.S, Zhang, F, Eisenmesser, E.Z. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of GeoCyp: A Thermophilic Cyclophilin with a Novel Substrate Binding Mechanism That Functions Efficiently at Low Temperatures.
Biochemistry, 54, 2015
|
|
2MW0
 
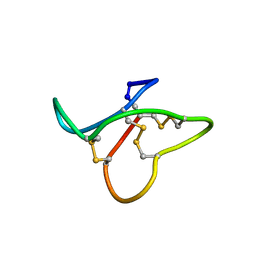 | |
2MW1
 
 | |
2MW2
 
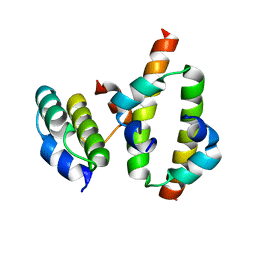 | | Hha-H-NS46 charge zipper complex | | Descriptor: | DNA-binding protein H-NS, Hemolysin expression-modulating protein Hha | | Authors: | Cordeiro, T.N, Garcia, J, Bernado, P, Millet, O, Pons, M. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Three-protein Charge Zipper Stabilizes a Complex Modulating Bacterial Gene Silencing.
J. Biol. Chem., 290, 2015
|
|
2MW3
 
 | | Solution NMR structure of the lasso peptide streptomonomicin | | Descriptor: | Lasso peptide | | Authors: | Tietz, J.I, Zhu, L, Mitchell, D.A, Metelev, M, Melby, J.O, Blair, P.M, Livnat, I, Severinov, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso Peptide from an understudied halophilic actinomycete.
Chem.Biol., 22, 2015
|
|
2MW4
 
 | | Tetramerization domain of the Ciona intestinalis p53/p73-b transcription factor protein | | Descriptor: | Transcription factor protein | | Authors: | Heering, J.P, Jonker, H.R.A, Loehr, F, Schwalbe, H, Doetsch, V. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigations of the p53/p73 homologs from the tunicate species Ciona intestinalis reveal the sequence requirements for the formation of a tetramerization domain.
Protein Sci., 25, 2016
|
|
2MW5
 
 | |
2MW6
 
 | | Structure of the bee venom toxin melittin with [(C5H5)Ru]+ fragment attached to the tryptophan residue | | Descriptor: | Melittin, [(1,2,3,4,5-eta)-cyclopentadienyl][(1,2,3,4,4a,8a-eta)-naphthalene]ruthenium(1+) | | Authors: | Perekalin, D.S, Pavlov, A.A, Novikov, V.V. | | Deposit date: | 2014-10-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Selective ruthenium labeling of the tryptophan residue in the bee venom Peptide melittin.
Chemistry, 21, 2015
|
|
2MW7
 
 | |
2MW8
 
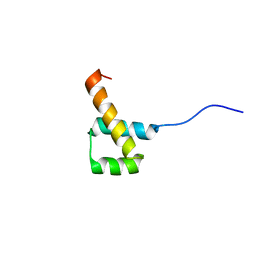 | |
2MW9
 
 | |
2MWA
 
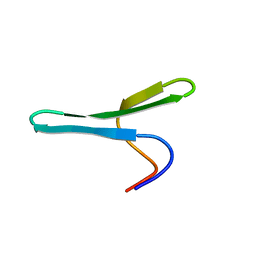 | |
2MWB
 
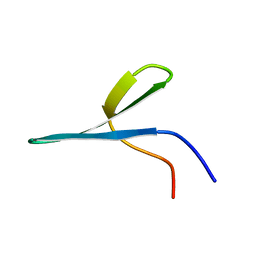 | | FBP28 WW2 mutant W457F | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MWC
 
 | |
2MWD
 
 | |
2MWE
 
 | | NMR structure of FBP28 WW2 mutant Y438R, L453A DNDC | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MWF
 
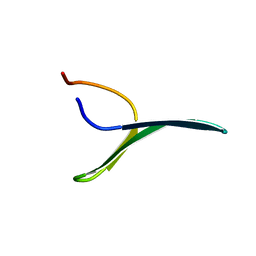 | |
2MWG
 
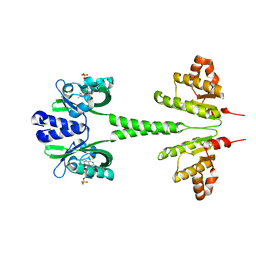 | |
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2MWI
 
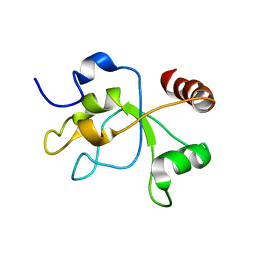 | | The structure of the carboxy-terminal domain of DNTTIP1 | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1 | | Authors: | Schwabe, J.W.R, Muskett, F.W, Itoh, T. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting.
Nucleic Acids Res., 43, 2015
|
|
2MWJ
 
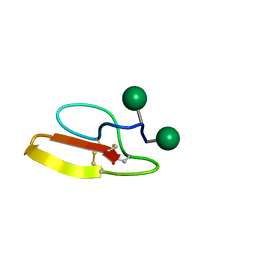 | | Solution structure of Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1 and Ser3 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2MWK
 
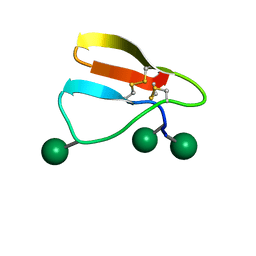 | | Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1, Ser3, and Ser14 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2MWL
 
 | |
