6YYO
 
 | | Structure of Cathepsin S in complex with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylsulfonylpiperazin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYP
 
 | | Structure of Cathepsin S in complex with Compound 2 | | Descriptor: | 1-(furan-2-ylmethyl)-5-(trifluoromethyl)benzimidazol-2-amine, ACETATE ION, Cathepsin S, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYQ
 
 | | Structure of Cathepsin S in complex with Compound 3 | | Descriptor: | (6~{R})-2-phenyl-5,6,7,8-tetrahydroquinazolin-6-amine, Cathepsin S | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYR
 
 | | Structure of Cathepsin S in complex with Compound 20b | | Descriptor: | (2~{R})-~{N}-(2-azanylideneethyl)-2-[2-(3-methyl-1,2-oxazol-5-yl)ethanoylamino]-3-(4-pyridin-2-ylpiperazin-1-yl)sulfonyl-propanamide, 1,2-ETHANEDIOL, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YZ3
 
 | | PqsR (MvfR) in complex with antagonist 19 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-[(2-hexyl-2,3-dihydro-1,3-thiazol-4-yl)methyl]quinazolin-4-one, LysR family transcriptional regulator | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
6YZ7
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
6YZD
 
 | | Crystal structure of the M295A variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase, SULFATE ION | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-05-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
6YZF
 
 | | Crystal structure of the M295Y variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase, GLU-HIS-SER, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-05-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
6YZY
 
 | | Crystal structure of the M295V variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase, SULFATE ION | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
6Z07
 
 | | PqsR (MvfR) in complex with antagonist 12 | | Descriptor: | 3-[(2-~{tert}-butyl-1,3-thiazol-4-yl)methyl]-6-chloranyl-quinazolin-4-one, GLYCEROL, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
6Z0U
 
 | |
6Z14
 
 | | Structure of Bifidobacterium bifidum GH20 beta-N-beta-N-acetylhexosaminidase E553Q variant in complex with 4MU-6SGlcNAc-derived oxazoline | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylhexosaminidase, NITRATE ION, ... | | Authors: | He, Y, Jin, Y, Rizkallah, P, Chen, P. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the GH20 beta-N-beta-N-acetylhexosaminidase from Bifidobacterium bifidum
To Be Published
|
|
6Z17
 
 | | PqsR (MvfR) in complex with antagonist 6 | | Descriptor: | 6-chloranyl-3-[(2-propan-2-yl-2,3-dihydro-1,3-thiazol-4-yl)methyl]quinazolin-4-one, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
6Z1P
 
 | |
6Z1Q
 
 | | MAP3K14 (NIK) in complex with DesF-3R/4076 | | Descriptor: | DesF-3R/4076, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Jacoby, E, van Vlijmen, H, Querolle, O, Stansfield, I, Meerpoel, L, Versele, M, Hynd, G, Attar, R. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | FEP+ calculations predict a stereochemical SAR switch for first-in-class indoline NIK inhibitors for multiple myeloma
Future Drug Discov, 2, 2020
|
|
6Z1T
 
 | | MAP3K14 (NIK) in complex with 4S/3694 | | Descriptor: | 4S/3694, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Jacoby, E, van Vlijmen, H, Querolle, O, Stansfield, I, Meerpoel, L, Versele, M, Hynd, G, Attar, R. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | FEP+ calculations predict a stereochemical SAR switch for first-in-class indoline NIK inhibitors for multiple myeloma
Future Drug Discov, 2, 2020
|
|
6Z2E
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone | | Descriptor: | (4~{S})-4-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[3-[2-[2-[2-[2-[5-[(3~{a}~{S},4~{R},6~{a}~{R})-2-oxidanylidene-3,3~{a},4,6~{a}-tetrahydro-1~{H}-thieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]butanoyl]amino]-3,3-dimethyl-butanoyl]amino]-4-methyl-pentanoyl]amino]-6-methylsulfonyl-hexanamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone
To Be Published
|
|
6Z2N
 
 | |
6Z33
 
 | |
6Z39
 
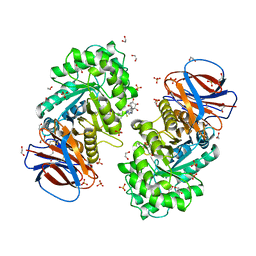 | | Structure of recombinant human beta-glucocerebrosidase in complex with BODIPY functionalised epoxide activity based probe | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[4-[4-[(12~{R})-2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9-trien-8-yl]butyl]-1,2,3-triazol-1-yl]methyl]-7-oxabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6Z3C
 
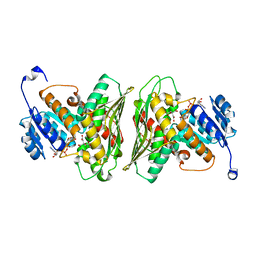 | | High resolution structure of RgNanOx | | Descriptor: | CITRATE ANION, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M.O. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6Z3I
 
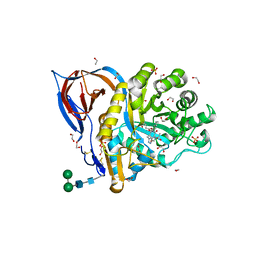 | | Structure of recombinant beta-glucocerebrosidase in complex with bifunctional cyclophellitol aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6Z3M
 
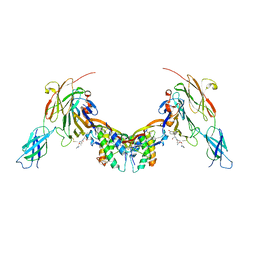 | | Repulsive Guidance Molecule B (RGMB) in complex with Growth Differentiation Factor 5 (GDF5) and Neogenin 1 (NEO1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 5, Neogenin, ... | | Authors: | Malinauskas, T, Peer, T.V, Bishop, B, Muller, T.D, Siebold, C. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (5.501 Å) | | Cite: | Repulsive guidance molecules lock growth differentiation factor 5 in an inhibitory complex.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z3N
 
 | |
6Z3O
 
 | |







