7I6M
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037755-001 (A71EV2A-x4986) | | Descriptor: | (3S)-N~1~-[2-(methylsulfanyl)-5-(2-oxopyrrolidin-1-yl)phenyl]piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6N
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037807-001 (A71EV2A-x4992) | | Descriptor: | (1S,4S,6R)-2-[(1-methyl-1H-pyrazol-4-yl)acetyl]-2-azabicyclo[4.1.0]heptane-4-carboxamide, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.385 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6O
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037765-001 (A71EV2A-x4994) | | Descriptor: | 1-[(1-methyl-1H-indazol-3-yl)acetyl]azetidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6P
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037779-001 (A71EV2A-x4998) | | Descriptor: | 1-{[4-(methylsulfanyl)thiophen-3-yl]acetyl}azetidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6Q
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037772-001 (A71EV2A-x5015) | | Descriptor: | (3S)-1-[(6-ethenylpyridin-2-yl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6R
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037768-001 (A71EV2A-x5016) | | Descriptor: | (3S)-1-[(2-ethenylpyridin-3-yl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6S
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037645-001 (A71EV2A-x5022) | | Descriptor: | (3S)-N~1~-[2-(methylsulfanyl)-5,6,7,8-tetrahydroquinolin-3-yl]piperidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6T
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037634-001 (A71EV2A-x5036) | | Descriptor: | (3S)-1-[(3-methoxyphenyl)methyl]-6-oxopiperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6U
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036740-001 (A71EV2A-x5039) | | Descriptor: | (3S)-1-{[4-(methylsulfanyl)pyrimidin-5-yl]acetyl}piperidine-3-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.456 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6V
 
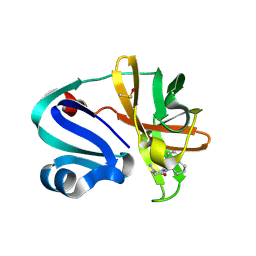 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037762-001 (A71EV2A-x5052) | | Descriptor: | (3S)-6-oxo-1-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6W
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037773-001 (A71EV2A-x5065) | | Descriptor: | (3S)-1-{2-[2-(methylsulfanyl)phenyl]ethyl}-5-oxopyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6X
 
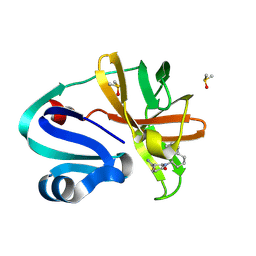 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037763-001 (A71EV2A-x5073) | | Descriptor: | (3S)-6-oxo-1-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6Y
 
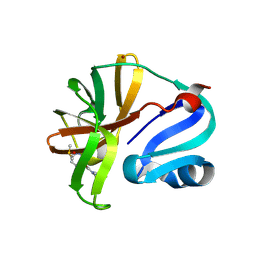 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037793-001 (A71EV2A-x5079) | | Descriptor: | (3S)-1-[(2-chloropyridin-3-yl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I6Z
 
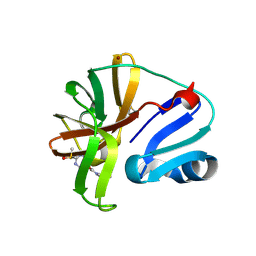 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037792-001 (A71EV2A-x5081) | | Descriptor: | (3S)-1-[(4-chloropyrimidin-5-yl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I70
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0037815-001 (A71EV2A-x5082) | | Descriptor: | (3S)-1-[(6-cyanopyridin-2-yl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I71
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038170-001 (A71EV2A-x5090) | | Descriptor: | (3S)-1-[(1-methyl-1H-pyrazol-4-yl)sulfamoyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I72
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038121-001 (A71EV2A-x5091) | | Descriptor: | (3S)-N~1~-methyl-N~1~-(3-methylphenyl)pyrrolidine-1,3-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I73
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038142-001 (A71EV2A-x5104) | | Descriptor: | (3S,5R)-5-methyl-1-{[2-(methylsulfanyl)phenyl]acetyl}piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I74
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038204-001 (A71EV2A-x5106) | | Descriptor: | (3S)-1-[(S)-(1-methyl-1H-imidazol-2-yl)(phenyl)methyl]-5-oxopyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I75
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038170-001 (A71EV2A-x5150) | | Descriptor: | (3S)-1-[(1-methyl-1H-pyrazol-4-yl)sulfamoyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I76
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038158-001 (A71EV2A-x5151) | | Descriptor: | (3S)-1-[(2,3-dihydro-1-benzothiophen-7-yl)acetyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I77
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038203-001 (A71EV2A-x5171) | | Descriptor: | (3R,5S)-1-{[2-(methylsulfanyl)phenyl]acetyl}piperidine-3,5-dicarboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I78
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038214-001 (A71EV2A-x5177) | | Descriptor: | (1S,4S,6R)-2-{[2-(methylsulfanyl)phenyl]acetyl}-2-azabicyclo[4.1.0]heptane-4-carboxamide, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I79
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038123-001 (A71EV2A-x5182) | | Descriptor: | (3S)-1-{[4-(methylsulfanyl)pyridin-3-yl]sulfamoyl}piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7I7A
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0038233-001 (A71EV2A-x5200) | | Descriptor: | (3S,5R)-5-methyl-1-{[2-(methylsulfanyl)phenyl]acetyl}piperidine-3-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
