7NYL
 
 | | Mutant H493A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | SH3 domain of JNK-interacting Protein 1 (JIP1), TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYM
 
 | | Mutant V517A - SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | HEXAETHYLENE GLYCOL, PHOSPHATE ION, SH3 domain of JNK-interacting Protein 1 (JIP1), ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYN
 
 | | Mutant Y526A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYO
 
 | | Mutant A541L of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, SH3 domain of JNK-interacting Protein 1 (JIP1), SULFATE ION, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZ2
 
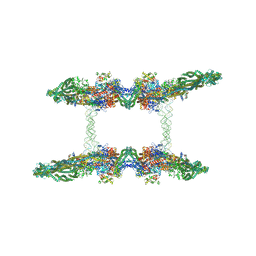 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ3
 
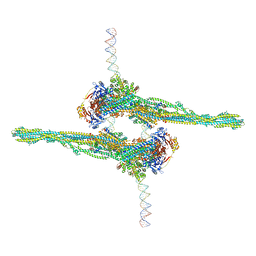 | | Cryo-EM structure of apposed MukBEF-MatP monomers on DNA | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
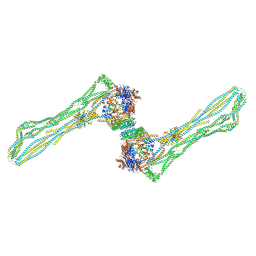 | | Cryo-EM structure of the MukBEF dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZB
 
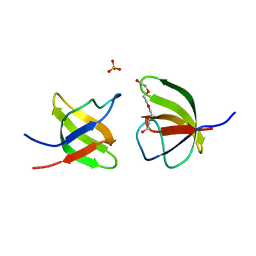 | | Mutant V517L of the SH3 domain of JNK-interacting protein 1 (JIP1) | | Descriptor: | PHOSPHATE ION, SH3 domain of JNK-interacting protein 1 (JIP1), TETRAETHYLENE GLYCOL | | Authors: | Perez, L.M, Ielasi, F.S, Jensen, M.R, Palencia, A. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZC
 
 | |
7NZD
 
 | | Fourth SH3 domain of POSH (Plenty of SH3 Domains protein) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase SH3RF1 | | Authors: | Palencia, A, Bessa, L.M, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZE
 
 | | Crystal structure of HLA-DR4 in complex with a human collagen type II peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen alpha-1(II) chain, GLYCEROL, ... | | Authors: | Ge, C, Dobritzsch, D, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZF
 
 | | Crystal structure of HLA-DR4 in complex with a mutated human collagen type II peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ge, C, Dobritzsch, D, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZH
 
 | | Crystal structure of HLA-DR4 in complex with a citrullinated cilp peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZQ
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7O00
 
 | | Crystal structure of HLA-DR4 in complex with a HSP70 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaperone protein DnaK, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2021-03-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7O0U
 
 | | Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0V
 
 | | Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0W
 
 | | Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0X
 
 | | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-28 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O18
 
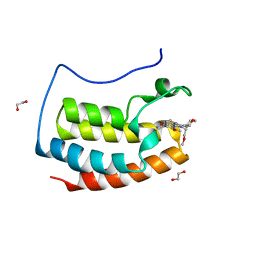 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH I-BET282 | | Descriptor: | (R)-4-(8-methoxy-1-(1-methoxypropan-2-yl)-2-(tetrahydro-2H-pyran-4-yl)-1H-imidazo[4,5-c]quinolin-7-yl)-3,5-dimethylisoxazole, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Novel Bromodomain and Extra Terminal Domain (BET) Protein Inhibitor, I-BET282E, Suitable for Clinical Progression.
J.Med.Chem., 64, 2021
|
|
7O19
 
 | | Cryo-EM structure of an Escherichia coli TnaC-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1A
 
 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1C
 
 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome-RF2 complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|






