+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1shx | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ephrin A5 ligand structure | |||||||||
 Components Components | Ephrin-A5 Ephrin A5 Ephrin A5 | |||||||||
 Keywords Keywords | HORMONE/GROWTH FACTOR / ephrin signaling / HORMONE-GROWTH FACTOR COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationneurotrophin TRKB receptor binding / EPH-Ephrin signaling / EPHA-mediated growth cone collapse / EPH-ephrin mediated repulsion of cells / negative regulation of substrate adhesion-dependent cell spreading / synaptic membrane adhesion /  regulation of cell-cell adhesion / collateral sprouting / cellular response to follicle-stimulating hormone stimulus / positive regulation of collateral sprouting ...neurotrophin TRKB receptor binding / EPH-Ephrin signaling / EPHA-mediated growth cone collapse / EPH-ephrin mediated repulsion of cells / negative regulation of substrate adhesion-dependent cell spreading / synaptic membrane adhesion / regulation of cell-cell adhesion / collateral sprouting / cellular response to follicle-stimulating hormone stimulus / positive regulation of collateral sprouting ...neurotrophin TRKB receptor binding / EPH-Ephrin signaling / EPHA-mediated growth cone collapse / EPH-ephrin mediated repulsion of cells / negative regulation of substrate adhesion-dependent cell spreading / synaptic membrane adhesion /  regulation of cell-cell adhesion / collateral sprouting / cellular response to follicle-stimulating hormone stimulus / positive regulation of collateral sprouting / regulation of insulin secretion involved in cellular response to glucose stimulus / regulation of cell-cell adhesion / collateral sprouting / cellular response to follicle-stimulating hormone stimulus / positive regulation of collateral sprouting / regulation of insulin secretion involved in cellular response to glucose stimulus /  chemorepellent activity / regulation of cell morphogenesis / retinal ganglion cell axon guidance / positive regulation of synapse assembly / chemorepellent activity / regulation of cell morphogenesis / retinal ganglion cell axon guidance / positive regulation of synapse assembly /  regulation of focal adhesion assembly / regulation of focal adhesion assembly /  regulation of GTPase activity / regulation of GTPase activity /  basement membrane / GABA-ergic synapse / ephrin receptor signaling pathway / regulation of microtubule cytoskeleton organization / cellular response to forskolin / basement membrane / GABA-ergic synapse / ephrin receptor signaling pathway / regulation of microtubule cytoskeleton organization / cellular response to forskolin /  ephrin receptor binding / ephrin receptor binding /  caveola / cell periphery / caveola / cell periphery /  axon guidance / regulation of actin cytoskeleton organization / axon guidance / regulation of actin cytoskeleton organization /  adherens junction / positive regulation of peptidyl-tyrosine phosphorylation / external side of plasma membrane / adherens junction / positive regulation of peptidyl-tyrosine phosphorylation / external side of plasma membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | |||||||||
 Authors Authors | Himanen, J.P. / Barton, W.A. / Nikolov, D.B. / Jeffrey, P.D. | |||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2005 Journal: J.Biol.Chem. / Year: 2005Title: Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3. Authors: Day, B. / To, C. / Himanen, J.P. / Smith, F.M. / Nikolov, D.B. / Boyd, A.W. / Lackmann, M. #1: Journal: Nat.Neurosci. / Year: 2004 Title: Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Authors: Himanen, J.P. / Chumley, M.J. / Lackmann, M. / Li, C. / Barton, W.A. / Jeffrey, P.D. / Vearing, C. / Geleick, D. / Feldheim, D.A. / Boyd, A.W. / Henkemeyer, M. / Nikolov, D.B. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1shx.cif.gz 1shx.cif.gz | 73.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1shx.ent.gz pdb1shx.ent.gz | 55.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1shx.json.gz 1shx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sh/1shx https://data.pdbj.org/pub/pdb/validation_reports/sh/1shx ftp://data.pdbj.org/pub/pdb/validation_reports/sh/1shx ftp://data.pdbj.org/pub/pdb/validation_reports/sh/1shx | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 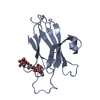
| ||||||||
| 2 | 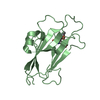
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  Ephrin A5 / EPH-related receptor tyrosine kinase ligand 7 / LERK-7 / AL-1 Ephrin A5 / EPH-related receptor tyrosine kinase ligand 7 / LERK-7 / AL-1Mass: 16320.332 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: EFNA5, EPLG7, LERK7, EPL7 / Cell line (production host): HEK293 / Organ (production host): kidney / Production host: Mus musculus (house mouse) / Gene: EFNA5, EPLG7, LERK7, EPL7 / Cell line (production host): HEK293 / Organ (production host): kidney / Production host:   Homo sapiens (human) / References: UniProt: O08543 Homo sapiens (human) / References: UniProt: O08543#2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2- ...2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose / triacetyl-beta-chitotriose | #3: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose |  / Mass: 424.401 Da / Num. of mol.: 1 / Mass: 424.401 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source #4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.26 Å3/Da / Density % sol: 45.58 % |
|---|---|
Crystal grow | Temperature: 278 K / Method: vapor diffusion, hanging drop / pH: 4.6 Details: PEG 3350, pH 4.6, VAPOR DIFFUSION, HANGING DROP, temperature 278K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X9A / Wavelength: 0.979 Å / Beamline: X9A / Wavelength: 0.979 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Dec 1, 2003 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.979 Å / Relative weight: 1 : 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 2→20 Å / Num. all: 17042 / Num. obs: 16487 / % possible obs: 91.3 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Net I/σ(I): 16.5 |
| Reflection shell | Resolution: 2→2.09 Å / Rmerge(I) obs: 0.056 / Num. unique all: 17958 / % possible all: 54.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: EprinB2 Resolution: 2.1→8 Å / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→8 Å
| ||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj



