+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1iv7 | ||||||
|---|---|---|---|---|---|---|---|
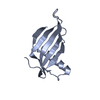
| Title | Crystal Structure of Single Chain Monellin | ||||||
 Components Components | Monellin | ||||||
 Keywords Keywords | PLANT PROTEIN / ALPHA+BETA | ||||||
| Function / homology |  Function and homology information Function and homology informationMonellin, A chain / Monellin, A chain superfamily / Monellin, B chain / : / Monellin / Monellin / Nuclear Transport Factor 2; Chain: A, - #10 / Cystatin superfamily / Nuclear Transport Factor 2; Chain: A, / Roll / Alpha Beta Similarity search - Domain/homology | ||||||
| Biological species |  Dioscoreophyllum cumminsii (serendipity berry) Dioscoreophyllum cumminsii (serendipity berry) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.82 Å MOLECULAR REPLACEMENT / Resolution: 1.82 Å | ||||||
 Authors Authors | Tamada, T. / Kato, Y. / Kuroki, R. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: The Effect of Single Chain Derivatization on the Structure and Stability of the Monellin Authors: Kato, Y. / Tamada, T. / Sone, H. / Iijima, H. / Kuroki, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1iv7.cif.gz 1iv7.cif.gz | 53.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1iv7.ent.gz pdb1iv7.ent.gz | 39 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1iv7.json.gz 1iv7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/iv/1iv7 https://data.pdbj.org/pub/pdb/validation_reports/iv/1iv7 ftp://data.pdbj.org/pub/pdb/validation_reports/iv/1iv7 ftp://data.pdbj.org/pub/pdb/validation_reports/iv/1iv7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1iv9C  1molS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 11287.853 Da / Num. of mol.: 2 / Mutation: E49N, N50E, E149N, N150E Source method: isolated from a genetically manipulated source Details: fusion protein concerning chain A comprise residues 1-50 (MONELLIN, CHAIN B) and 52-96 (MONELLIN, CHAIN A), Including glysine linker. fusion protein concerning chain B comprise residues 101- ...Details: fusion protein concerning chain A comprise residues 1-50 (MONELLIN, CHAIN B) and 52-96 (MONELLIN, CHAIN A), Including glysine linker. fusion protein concerning chain B comprise residues 101-150 (MONELLIN, CHAIN B) and 152-196 (MONELLIN, CHAIN A), Including glysine linker. Source: (gene. exp.)  Dioscoreophyllum cumminsii (serendipity berry) Dioscoreophyllum cumminsii (serendipity berry)Plasmid: pST6311 / Species (production host): Escherichia coli Production host:  Strain (production host): W3110 / References: UniProt: P02882, UniProt: P02881 #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.26 Å3/Da / Density % sol: 45.51 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: microbatch / pH: 7.2 Details: PEG8000, potassium phosphate, pH 7.2, micro batch, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 277 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200H / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200H / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: Mar 19, 1998 |
| Radiation | Monochromator: MIRRORS / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→60 Å / Num. all: 18113 / Num. obs: 17026 / % possible obs: 94 % / Observed criterion σ(I): 0 / Rmerge(I) obs: 0.067 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1MOL Resolution: 1.82→15 Å / Cor.coef. Fo:Fc: 0.959 / Cor.coef. Fo:Fc free: 0.941 / SU B: 4.762 / SU ML: 0.136 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.162 / ESU R Free: 0.149 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.272 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.82→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.824→1.871 Å / Total num. of bins used: 20 /
|
 Movie
Movie Controller
Controller











 PDBj
PDBj

