[English] 日本語
 Yorodumi
Yorodumi- SASDBQ9: Human Hsp90 co-chaperone Cdc37 (CD37) in complex with fibroblast ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDBQ9 |
|---|---|
 Sample Sample | Human Hsp90 co-chaperone Cdc37 (CD37) in complex with fibroblast growth factor receptor 3
|
| Function / homology |  Function and homology information Function and homology informationt(4;14) translocations of FGFR3 / negative regulation of developmental growth / fibroblast growth factor receptor apoptotic signaling pathway / Signaling by FGFR3 fusions in cancer / bone maturation / regulation of type II interferon-mediated signaling pathway / HSP90-CDC37 chaperone complex / chondrocyte proliferation / endochondral bone growth / FGFR3b ligand binding and activation ...t(4;14) translocations of FGFR3 / negative regulation of developmental growth / fibroblast growth factor receptor apoptotic signaling pathway / Signaling by FGFR3 fusions in cancer / bone maturation / regulation of type II interferon-mediated signaling pathway / HSP90-CDC37 chaperone complex / chondrocyte proliferation / endochondral bone growth / FGFR3b ligand binding and activation / Signaling by activated point mutants of FGFR3 / FGFR3c ligand binding and activation / positive regulation of phospholipase activity / Phospholipase C-mediated cascade; FGFR3 / endochondral ossification / fibroblast growth factor receptor activity / positive regulation of type 2 mitophagy / regulation of cyclin-dependent protein serine/threonine kinase activity / protein kinase regulator activity / bone morphogenesis / protein folding chaperone complex / positive regulation of tyrosine phosphorylation of STAT protein / PI-3K cascade:FGFR3 / post-transcriptional regulation of gene expression / Drug-mediated inhibition of ERBB2 signaling / Resistance of ERBB2 KD mutants to trastuzumab / Resistance of ERBB2 KD mutants to sapitinib / Resistance of ERBB2 KD mutants to tesevatinib / Resistance of ERBB2 KD mutants to neratinib / Resistance of ERBB2 KD mutants to osimertinib / Resistance of ERBB2 KD mutants to afatinib / Resistance of ERBB2 KD mutants to AEE788 / Resistance of ERBB2 KD mutants to lapatinib / Drug resistance in ERBB2 TMD/JMD mutants / bone mineralization / regulation of type I interferon-mediated signaling pathway / fibroblast growth factor binding / PI3K Cascade / fibroblast growth factor receptor signaling pathway / chondrocyte differentiation / cell surface receptor signaling pathway via JAK-STAT / RHOBTB2 GTPase cycle / SHC-mediated cascade:FGFR3 / protein targeting / transport vesicle / FRS-mediated FGFR3 signaling / heat shock protein binding / Signaling by FGFR3 in disease / Signaling by ERBB2 / Constitutive Signaling by Overexpressed ERBB2 / skeletal system development / Negative regulation of FGFR3 signaling / Signaling by ERBB2 TMD/JMD mutants / Hsp90 protein binding / Constitutive Signaling by EGFRvIII / receptor protein-tyrosine kinase / Signaling by ERBB2 ECD mutants / Signaling by ERBB2 KD Mutants / Regulation of necroptotic cell death / kinase binding / Downregulation of ERBB2 signaling / Constitutive Signaling by Aberrant PI3K in Cancer / unfolded protein binding / protein folding / PIP3 activates AKT signaling / cell-cell signaling / MAPK cascade / Constitutive Signaling by Ligand-Responsive EGFR Cancer Variants / protein-folding chaperone binding / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / RAF/MAP kinase cascade / protein tyrosine kinase activity / scaffold protein binding / positive regulation of ERK1 and ERK2 cascade / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / receptor complex / positive regulation of MAPK cascade / protein stabilization / positive regulation of cell population proliferation / protein kinase binding / cell surface / endoplasmic reticulum / Golgi apparatus / extracellular exosome / extracellular region / ATP binding / identical protein binding / plasma membrane / cytosol / cytoplasm Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2018 Mar Date: 2018 MarTitle: Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System Authors: Bunney T / Inglis A / Sanfelice D / Farrell B / Kerr C / Thompson G / Masson G / Thiyagarajan N / Svergun D / Williams R / Breeze A |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
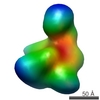
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDBQ9 SASDBQ9 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #1132 |  Type: mix / Software: (ATSAS2.7.2) / Radius of dummy atoms: 3.50 A / Symmetry: P1 / Chi-square value: 2.17  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #1133 |  Type: dummy / Software: (ATSAS2.7.2) / Radius of dummy atoms: 3.00 A / Symmetry: P1 / Chi-square value: 1.443  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Human Hsp90 co-chaperone Cdc37 (CD37) in complex with fibroblast growth factor receptor 3 Specimen concentration: 1.00-3.80 / Entity id: 585 / 589 |
|---|---|
| Buffer | Name: 25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP pH: 8 |
| Entity #585 | Name: CDC37 / Type: protein / Description: Hsp90 co-chaperone Cdc37 / Formula weight: 44.426 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q16543 Sequence: MVDYSVWDHI EVSDDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEGGKAE LERLQAEAQQ LRKEERSWEQ KLEEMRKKEK SMPWNVDTLS KDGFSKSMVN TKPEKTEEDS EEVREQKHKT FVEKYEKQIK HFGMLRRWDD ...Sequence: MVDYSVWDHI EVSDDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEGGKAE LERLQAEAQQ LRKEERSWEQ KLEEMRKKEK SMPWNVDTLS KDGFSKSMVN TKPEKTEEDS EEVREQKHKT FVEKYEKQIK HFGMLRRWDD SQKYLSDNVH LVCEETANYL VIWCIDLEVE EKCALMEQVA HQTIVMQFIL ELAKSLKVDP RACFRQFFTK IKTADRQYME GFNDELEAFK ERVRGRAKLR IEKAMKEYEE EERKKRLGPG GLDPVEVYES LPEELQKCFD VKDVQMLQDA ISKMDPTDAK YHMQRCIDSG LWVPNSKASE AKEGEEAGPG DPLLEAVPKT GDEKDGSV |
| Entity #589 | Name: FGFR3 / Type: protein / Description: Fibroblast growth factor receptor 3 / Formula weight: 35.478 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: P22607 Sequence: SELELPADPK WELSRARLTL GKPLGEGAFG QVVMAEAIGI DKDRAAKPVT VAVKMLKDDA TDKDLSDLVS EMEMMKMIGK HKNFINLLGA CTQGGPLYVL VEYAAKGNLR EFLRARRPPG LDYSFDTSKP PEEQLTFKDL VSCAYQVARG MEYLASQKCI HRDLAARNVL ...Sequence: SELELPADPK WELSRARLTL GKPLGEGAFG QVVMAEAIGI DKDRAAKPVT VAVKMLKDDA TDKDLSDLVS EMEMMKMIGK HKNFINLLGA CTQGGPLYVL VEYAAKGNLR EFLRARRPPG LDYSFDTSKP PEEQLTFKDL VSCAYQVARG MEYLASQKCI HRDLAARNVL VTEDNVMKIA DFGLARDVHN LDYYKKTTNG RLPVKWMAPE ALFDRVYTHQ SDVWSFGVLL WEIFTLGGSP YPGIPVEELF KLLKEGHRMD KPANCTHDLY MIMRECWHAA PSQRPTFKQL VEDLDRVLTV TSTDEYLDLS APFE |
-Experimental information
| Beam | Instrument name: PETRA III P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||||||||||||||||||||
| Scan | Measurement date: Jul 10, 2016 / Storage temperature: 20 °C / Cell temperature: 20 °C / Exposure time: 0.045 sec. / Number of frames: 20 / Unit: 1/nm /
| ||||||||||||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller